CREST Teaching and Research Symposium – April 28, 2012
The second year of the CREST Project included several new modeling projects and expansions of existing modeling projects to provide a more complete molecular story of protein function, as well as the development of several instructional materials.
A combination of CREST educators and undergraduates provided progress reports on the modeling and instructional materials development projects at their institutions, and a few educators were able to provide a framework for assessment of the impact of the instructional materials in the classroom. (Piloting of several of the materials began in spring of 2012, and the symposium was held before the completion of the spring term, so final data was not available for presentation at the symposium.)
Project Updates
- Overview of CREST Program
- Mount Mary College
- Jmol Tutorials
- Alverno College
- Concordia University Wisconsin
- Marquette University
- University of Wisconsin-Madison
- Clickable Molecular Landscape
- Jmol Tutorials
- University of Wisconsin-Milwaukee
- Stony Brook University – assessment
- Tina M. Jeselun
- Sara. I. Klosiewski
- Phaya Lem
- Jessica. L. Weis
- Carl Ball, Ph.D.
- Howard Jacob, Ph.D., Human Medical Genetics Center, Medical College of Wisconsin
- inw9.pdb
- Danielle Burke
- Christina Evans
- Anne Grulke
- Ann McDonald, Ph.D.
- Amy Hudson, Ph.D.; Medical College of Wisconsin, Milwaukee WI
- 1im3.pdb
- Powerpoint
- Poster (The Concordia University Wisconsin 2011-2012 CREST Team presented their poster at the American Society of Microbiology Conference for Undergraduate Educators in San Mateo CA.)
- Jessica Benson
- Amy Ramirez
- Nerissa Seward
- Colleen Conway, Ph.D.
- Jung-Ja Kim, Ph.D.; Medical College of Wisconsin, Milwaukee WI
- 1t9g.pdb
- 1udy.pdb
- 2ait.pdb
- Ann Cassany Anselme
- Amanda Morris
- Megan Nemeth
- Vasty Souffrant
- Audrey Shor, Ph.D., M.P.H.
- Nikita Patel, Ph.D., James A Haley Veterans Association, Tampa, FL
- 1bdy.pdb
- Sean Lastorka
- Issys Salazar
- Joseph Zundell
- Audrey Shor, Ph.D., M.P.H.
- Tomar Ghansah, Ph.D., University of South Florida College of Medicine, Tampa, FL
- 2ysx.pdb
- Joseph Johnston
- Bryan Landrie
- Anne Marie Wannamaker
- Steven Forst, Ph.D.
- 3e0d.pdb
Alverno College
Interaction Between Caspase-9 and Mutated Residues on the Third Domain (BIR3) of XIAP

Students:
Faculty Advisor:
Research Mentor:

PDB file:
Presentation:
Abstract:
The BIR3 (baculovirus IAP) domain of XIAP binds to caspase-9 to form a complex that inhibits apoptosis. BIR3 prevents the dimerization of caspase-9 by blocking its active site. Four specific amino acids in BIR3 are important to apoptosis inhibition by the BIR3/caspase-9 complex. BIR3 mutants still form the complex in vitro, but allow caspase-9 to homodimerize. The structure and size of each conserved residue in BIR3 are predicted to affect the tight binding complex with caspase-9. In vitro studies suggest that the interaction between mutated BIR3 and caspase-9 would affect the complex to deregulate caspase-9. This could allow for an increase in apoptosis, but it is uncertain how this would impact living organisms, which raises questions for further research.
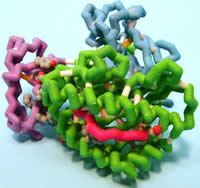
Concordia University Wisconsin
Human Cytomegalovirus Protein U2 Bound to HLA-A2/tax
Students:
Faculty Advisor:
Research Mentor:
PDB file:

Presentation:
Abstract:
Viruses are parasitic microorganisms that invade cells to replicate themselves. Once replicated, many viruses kill the host cell so their progeny can invade other cells, repeating the replication process and ensuring their survival. Some viruses have developed mechanisms to avoid the host immune system and can therefore establish latent infections.Cells in the human immune system recognize the presence of infectious microorganisms and eliminate them. Recognition of viral-infected cells is mediated by cell surface proteins called class I major histocompatibility (MHC) proteins. Class I MHC proteins specifically bind small portions of viral proteins (peptides) on the surface of infected cells and present them to cytotoxic T lymphocytes (CTL), targeting the infected cell for destruction by the CTL. Human cytomegalovirus (HCMV) is a herpes virus that can establish latent infections, due to a viral protein called unique short 2 (US2) glycoprotein. X-ray crystallography of US2 identified the presence of an immunoglobulin-fold (Ig-fold), composed of two β-sheets where the β-strands run anti-parallel. The Ig-fold appears to play a role in evading the host’s immune system; however, the mechanism is not yet known. When US2 binds to Class I MHC in the endoplasmic reticulum (ER), the molecules are sent to the cytoplasm for degradation. Immunoprecipitation studies suggest that a protein called signal peptide peptidase (SPP) may be an essential factor in the degradation or presentation of the viral peptide to Class I MHC. Therefore, US2 helps establish latent infections with HCMV by inhibiting class I MHC presentation of viral peptides to CTL.
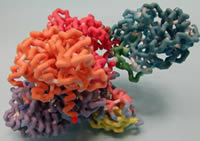
Mount Mary College
Medium Chain Acyl-CoA Dehydrogenase (MCAD) with Associated Electron Transfer Flavoprotein (ETF)
Students:
Faculty Advisor:
Research Mentor:

PDB files:
Presentation:
Abstract:
SIDS is a somewhat common diagnose in infant deaths and it was discovered that some of these cases were due to defects in the medium chain acyl CoA dehydrogenase (MCAD)enzyme. If this had been known earlier, the MCAD deaths may have been prevented. This is a common defect in people of northern European descent. MCAD is the first enzyme in the mitochondrial beta oxidation spiral. This enzyme removes two hydrogens and creates a carbon-carbon double bond in the substrate. This enzyme only works on fatty acyl-CoA substrates of 4 to 12 carbons. There is a family of 9 of these acyl-CoA dehydrogenases enzymes that do the same thing except on different lengths of carbon chain. The most common are short chain acyl-CoA dehydrogenase, long chain acyl-CoA dehydrogenase, and very long chain acyl-CoA dehydrogenase. The other enzymes in the other steps of the beta oxidation of fatty acids also have a family of enzymes that work on different chain lengths of substrate. This enzyme is a homotetramer with 4 identical units in the enzyme and an ETF (electron transport flavoprotein) also attached to one subunit. Each subunit has an FAD (flavin adenine dinucleotide) molecule that is the electron transporter for electrons from the substrate (acyl-CoA) to ETF and eventually to the ETC. These structures are fairly conserved in the other acyl-CoA dehydrogenases and MCAD was often used as the model for others.
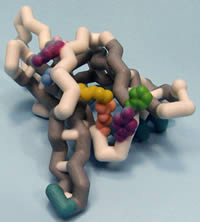
Saint Leo University
Protein Kinase C δI C2 Domain
Students:
Faculty Advisor:
Research Mentor:
PDB file:
Presentation:

Abstract:
Protein Kinase C delta (PKCδ) is a newly characterized member of the PKC family, a group of 11 isozymes classified as serine/threonine protein kinases. This family plays a critical role in regulating several signal transduction pathways which modulate cellular events. The general structure is divided in three regions: a catalytic domain, regulatory domain, and N terminus. PKCδ is a complex molecule comprised of 9 isoforms observed in human (I/VIII), rat (I/III) and mouse (I/II/IV-VII/IX). Function differs with cellular location and includes promoting cell survival, growth and development, tumor suppression and apoptosis. Alternative expression of the PKCδ isoforms has been demonstrated to regulate cellular differentiation during neurogenesis (PKCδII) and initiation of apoptosis (PKCδI). Unlike conventional PKCs, PKCδ is activated by diacylglycerol and phospholipid in a Ca2+-independent manner. The regulatory domain of PKCδ includes C1 and C2 motifs separated by a hinge region (V3), which regulates protein activation. This model emphasizes the C2 domain of PKCδI, which targets the protein for expression at the membrane. Three unique residues of tyrosine are significant for function: Tyr12 replaces a highly conserved alanine or glycine and is involved in stabilizing a hydrophobic core of the protein; Tyr52 is an essential phospshopeptide regulatory site; and Tyr64 regulates nuclear localization of the protein and induction of apoptosis. Residues essential for docking binding partners to this signaling protein include Thr50, a potential docking site for Src Homology 2 domains, and a phosphopeptide binding site(Lys48, His62 and Arg67), which constitutes a hydrophilic motif that initiates interactions with binding partners.
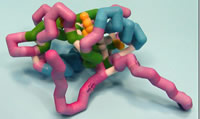
Saint Leo University
Signaling Inositol Polyphosphate Phosphatase I Src-Homology 2 Domain SHIP-1 SH2 Domain
Students:
Faculty Advisor:
Research Mentor:

PDB file:
Presentation:
Abstract:
SH2 containing Inositol 5-Phosphatase (SHIP-1) is a serine/tyrosine protein phosphatase expressed in blood cells that negatively regulates the Phosphoiniositide-3-Kinase (PI3K) pathway. SHIP-1 plays an essential role in immune system maintenance by regulating proliferation, differentiation, survival/apoptosis, and B-cell antigen receptor signaling. Recent evidence has suggested that SHIP-1 may even act as a tumor suppressor, while other data suggests it promotes tumor progression. Most recently, SHIP-1 expression has been demonstrated to be down regulated in a mouse model of pancreatic cancer. The structure of SHIP-1 contains a Src homology 2 (SH2) domain, a proline rich SH3 domain (involved in recruitment to the plasma membrane), and a catalytic domain (C2, observed in many protein kinases). The SH2 domain provides essential scaffolding for anchoring to protein complexes essential for transducing SHIP-1 initiated signals via phospho-tyrosine recognition. The serine residues 30 and 36 are essential for anchoring SHIP-1 to signaling partners. When mutated, SHIP-1 was unable dephosphorylate phosphatidylinositol (3,4,5) triphosphate and inhibit the PI3K pathway. Two arginine residues (34 and 41) are also essential for the inflammatory response mediated by SHIP-1. When mutated at Arg41, oxidative stress induced responses in leukemic cells are inhibited. SHIP-1 with mutated Arg34 exhibit diminished interaction with molecules essential for red blood cell production. Further evaluation of SHIP-1 regulation of the pro-survival PI3K pathway will provide insight for better understanding the phenotype observed in SHIP-1 mutational and knockout studies. In examining SHIP-1, medicinal compounds can be produced to stimulate activity.
University of Wisconsin – Milwaukee
Thermus aquaticus DNA Polymerase III, α Subunit
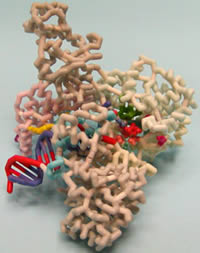
Students:
Faculty Advisor:
PDB file:
Presentation:
Abstract:

DNA polymerases function in the replication and repair of chromosomes. Bacterial chromosome replication is performed by the replisome, a multi-subunit holoenzyme complex, comprised of a core polymerase complex (α, ε, θ), a clamp loader complex (γ, τ2, δ, δ, χ, ψ), and a β-sliding clamp. A crystal structure of the DNA polymerase III α-subunit (PolIIIα) of Thermus aquaticus (Taq) with a template DNA fragment and a deoxynucleotide has been recently determined. In the present project a physical model of PolIIIα was created. The Taq α-subunit has the similar “hand-like” structure of PolIIIα of E.coli and is composed of six subdomains: palm, fingers, thumb, histidinol phosphatase (PHP), β-binding, and a C-terminal domain (CTD). The catalytic palm domain is similar to the eukaryotic DNA repair polymerase, Polβ, and not eukaryotic replicative DNA polymerases. Three highly conserved aspartate residues (D463, D465, D618) within the palm domain coordinate divalent metals involved in catalysis. A conserved glycine-serine motif (G425, S426) and four conserved arginine residues (R452, 458, 766, 767) participate in incoming nucleotide binding. A lysine residue (K616) near the catalytic site positions the 3’-terminal phosphate of the primer strand for dNTP addition. Incoming nucleotides undergo deoxyribose selection and pre-catalytic positioning in an entry channel through the finger domain. Upon binding DNA, the α-subunit undergoes a conformational shift allowing interactions with the sugar-phosphate backbone. Several subdomains contribute to forming a DNA binding pocket. Interaction of a loop from the β-binding site with a cleft within the β-clamp supports a model for polymerase switching.