



This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
1. Recognize that molecular interaction between enzyme & substrate lowers the activation energy required for a thermodynamically feasible reaction to occur.2. Recognize that primary through tertiary (and in some cases quaternary) structure determines the catalytic & allosteric functional domains of enzymes.3. Understand that catalytic domain structure & orientation determines enzyme-substrate binding.4. Understand how enzyme functional domain structure can be changed by addition of chemical groups like phosphates, and how these enzyme structural changes affect enzymatic activity.
Remember to interact with the model. Click on the molecule and drag your mouse to spin the molecule. Scroll your mouse to zoom in and out. When you pause your mouse over a kink, a small pop up window will appear with a three-letter identifier for that amino acid (e.g., PRO for proline).
This section reviews some important terminology. Refer back to this section as needed throughout the tutorial as you learn about the different proteins (use the 'Content' bar on the top left of your screen to jump to different sections of this tutorial).
Backbone- this representation shows the peptide chain only, without the side chains (also called residues or R groups). Each kink on a chain represents the central α carbon of one amino acid. This is shown in grey in the model to the right.
Ball and stick- this representation is used to shows particular amino acid side chains. Each ball and stick color on a sidechain represents a different atom: white for hydrogen, black for carbon, blue for nitrogen, red for oxygen, yellow for sulfur. You do NOT have to memorize these colors, but note that the specific atoms that compose these side chains determine the properties of the amino acid (polar, nonpolar, negatively charged, etc.) and thus how the protein interacts with other biomolecules!
The job of an enzyme is to catalyze a reaction. That is, enzymes increase the rate of a reaction by lowering the activation energy, thus allowing a thermodynamically feasible reaction to occur. Most enzymes are proteins, and as such, their function depends on primary through quaternary protein structure just as you learned in the previous tutorial. As you know, interactions between residues (R groups) result in secondary protein structures like alpha helices and beta sheets. Tertiary structure (protein folding) allows for the formation of functional domains. In the case of enzymes, one key functional domain is the active site, where substrate binding and then catalysis occur.
Enzymatic activity is often regulated by changing enzymatic structure, sometimes just by adding simple chemical groups like phosphates. We will use the enzyme glycogen phosphorylase as a case study to better understand how enzymatic activity can be regulated through structural changes.
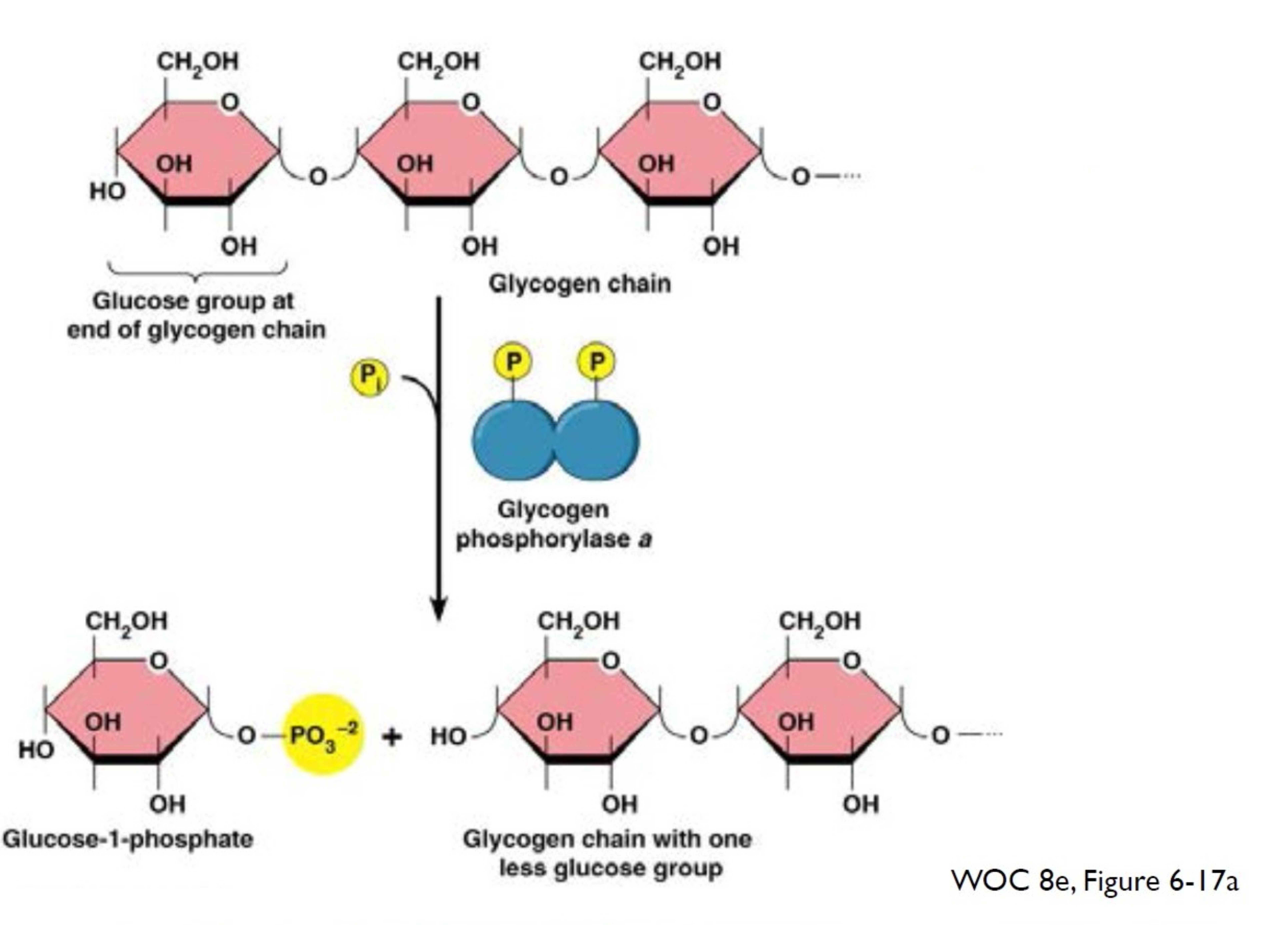
Glycogen is made up of long chains of glucose molecules that can be easily broken down when energy is needed. Glycogen phosphorylase is a dimeric enzyme (made of two monomers) in muscle cells that catalyzes the cleavage of glucose from the glycogen chain. Glycogen phosphorylase removes glucose units as glucose-1-phosphate. Ultimately, the released glucose-1-phosphate can enter glycolysis; the action of glycogen phosphorylase is therefore important for glycogen metabolism (see image below).
This glycogen phosphorylase enzyme can exist in either an active, phosphorylated form or an inactive, unphosphorylated form. Only the active form of glycogen phosphorylase can cleave individual glucose units from glycogen chains (see image below).
Recall that proteins adopt secondary structure based on their primary structure. Hydrogen bonding between specific amino acids determines whether α-helices & β-sheets will form. Click the button below to see the secondary structure(s) of one glycogen phosphorylase protein chain.
Glycogen phosphorylase secondary structureWhat are the red structures? Where do hydrogen bonds form to make these structures?
What are the yellow structures? Where do hydrogen bonds form to make these structures?
Primary and secondary structure helps to determine tertiary structure (protein folding) and allow for the formation of functional domains. In the case of glycogen phosphorylase, proper folding allows for the formation of a glycogen-binding active site. The active site includes a cofactor pyridoxal-5'-phosphate (PLP), which covalently binds the substrate, glycogen. PLP is a cofactor for many enzymes involved in metabolism. Click the button below to see one glycogen phosphorylase protein (monomer).
Glycogen phosphorylase active siteIn this model, the backbones of regions involved in inhibition of the active site are colored red and the regions involved in catalysis are colored green. The cofactor PLP is displayed in ball and stick and colored in light CPK. The PLP position indicates the active site location. The N-terminus is blue and C-terminus is magenta.
Which residues are closer to the substrate binding site: those involved in inhibition or those involved in catalysis of the glycogen substrate?
Zoom out to view the entire protein. How large is the active site compared to the rest of the protein?
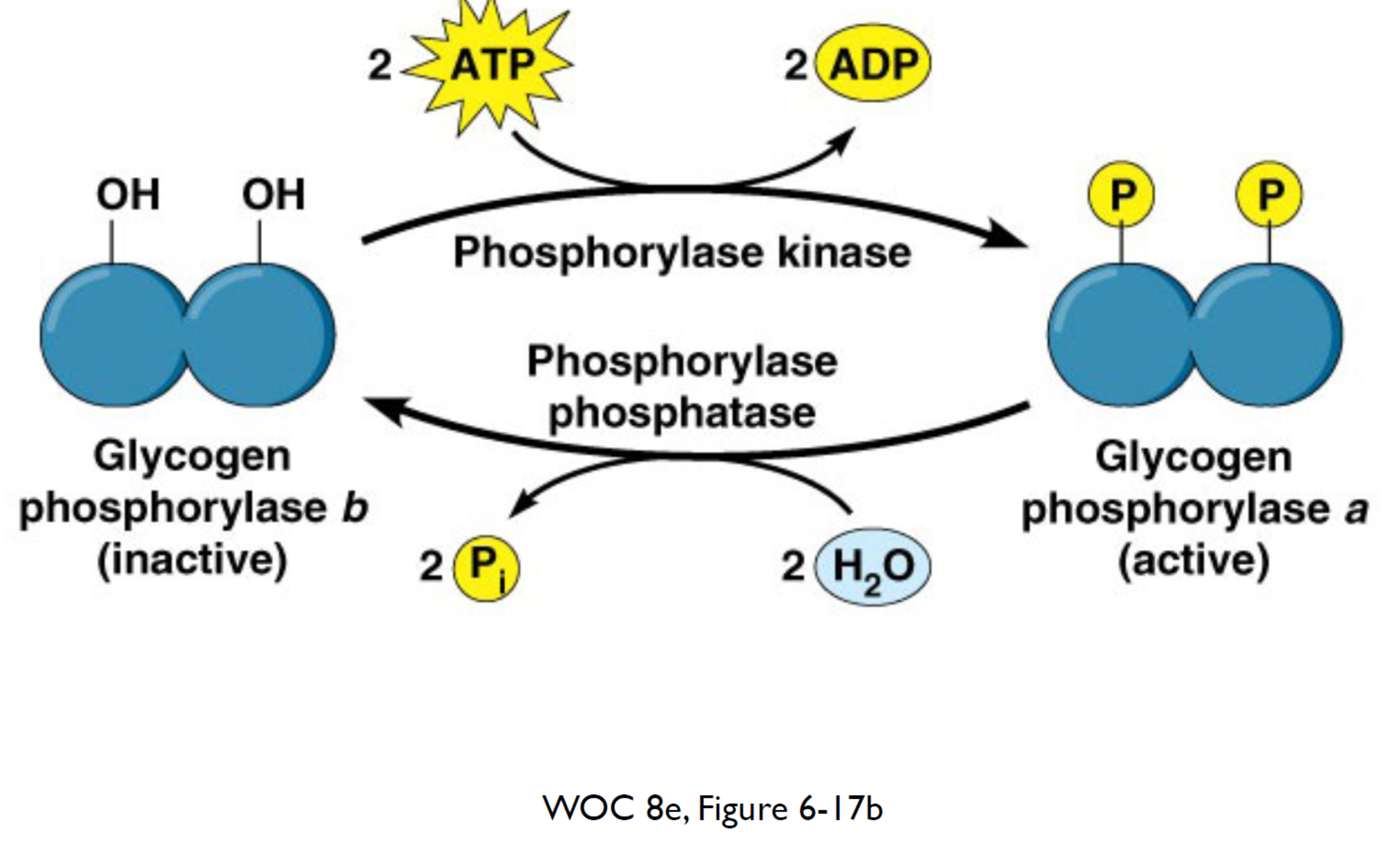
Enzymes typically have higher activity when they are in an 'open' state (and therefore available for substrate binding) compared to a 'closed' state (and therefore unavailable for substrate binding). In the case of glycogen phosphorylase, the shift between the two shapes is controlled by phosphorylation of serine 14 on each monomer, which induces the 'open' state and increases enzymatic activity. Locate serine 14 on the model shown to the right.
Where is the serine located in comparison to the active site?
This type of regulation (phosphorylation) is one type of covalent modification. Covalent modifications alter an enzyme's activity by the addition or removal of chemical groups, which alter the interactions of amino acid residues within the enzyme. In addition to phosphorylation, other common modifications include addition of methyl groups and acetyl groups. Each of these chemical groups has different properties that will alter the protein's conformation in a different way.
What might be a key property of a phosphate group that allows it to alter the conformation of a protein?
A phosphate group has a negative charge. Serine 14 is located near the N-terminus, which contains many negatively charged residues. When a phosphate group is added to serine 14, it repels the negatively charged amino acids in the N-terminus and the N-terminus moves. This movement results in a significant conformational change throughout the protein, which causes the active site to adopt the 'open' state, thus increasing enzymatic activity.
Without the presence of the phosphate groups on the two serines (remember that glycogen phosphorylase is a dimer), glycogen phosphorylase remains in a 'closed' state, which is less accessible for substrate binding. Click here glycogenPhosphorylase.html to open a new window that compares the open/active and closed/inactive glycogen phosphorylase states. Examine the active site as well as the position of the N-terminus in both states. Note that this will take 10-20 seconds to load. Remember to scroll your mouse to zoom in and out.
What do you notice about the differences in glycogen phosphorylase structure in the phosphorylated vs. unphosphorylated states? Is the active site truly 'closed' when it is unphosphorylated?
Covalent modifications such as phosphorylation involve a somewhat permanent alteration to a protein. These alterations can only be reversed through the action of a second enzyme. In the case of glycogen phosphorylase, the open/active phosphorylated form is returned to a closed/inactive form when the serine 14 phosphate groups are removed by the enzyme phosphorylase phosphatase (see WOC figure 6-17b below).
In contrast, allosteric regulation typically involves a reversible interaction between a small organic molecule and a regulatory domain within an enzyme. Such interactions are non-covalent. Glycogen phosphorylase is regulated allosterically by AMP (adenine monophosphate), which binds to a site on the opposite side of the protein from the active site, causing the same shift to the active, open conformation. Remember that AMP is a product of ATP breakdown.
Why is it biologically useful for AMP to activate glycogen phosphorylase?
In order to function, glycogen phosphorylase must adopt quaternary structure (the interaction of two or more protein subunits). A single glycogen phosphorylase protein (monomer) must bind to another glycogen phosphorylase monomer to be functional. Click the button below to see the functional glycogen phosphorylase dimer.
active glycogen phosphorylase dimerHow many chains of glycogen can be bound in this model?
Glycogen phosphorylase activity is also regulated by its product (glucose-6 phosphate). Cleavage of glycogen to mobilize more glucose is unnecessary when glucose concentration is high. It turns out that glucose-6 phosphate molecules can bind to the active site of glycogen phosphorylase to inhibit the enzyme. This is an excellent example of negative feedback, or end product inhibition.
What might happen to effective Km if glucose-6 phosphate is bound?