



This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
Students will be able to recognize/ predict/ graph/hypothesize/ draw a diagram/ design an experiment to demonstrate their understanding of how gene expression is regulated by:1. transcription factor structure.2. transcription factor location within the cell.3. dynamic interactions between transcription factors, regulatory proteins and DNA.
There is some important terminology to go over before you begin. Refer back to this section as needed throughout the tutorial as you learn about the different proteins (use the 'Contents' bar on the top left of your screen to jump to different sections of this tutorial).
Backbone- this representation shows the peptide chain only, without the side chains (also called residues or R groups). Each kink on a chain represents the central α carbon of one amino acid.
Ball and stick- this representation shows particular amino acid side chains. Each ball and stick color on a sidechain represents a different atom: white for hydrogen, black for carbon, blue for nitrogen, red for oxygen, yellow for sulfur. You do NOT have to memorize these colors, but note that the specific atoms that compose these side chains determine the properties of the amino acid (polar, nonpolar, negatively charged, etc.) and thus how the protein interacts with other biomolecules!
Biomolecular structure, location, and dynamic interactions with other molecules affect key cellular functions like gene expression (see Fig. 1). Gene expression in a particular cell at a particular time depends on many factors, which allows for dynamic regulation in response to extracellular signals.
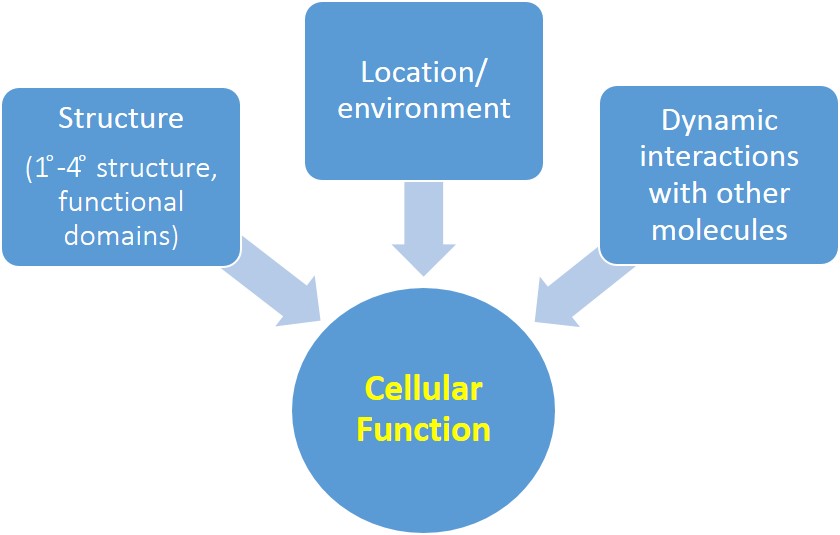
What is the role of transcription factors in the regulation of gene expression? How is the activity of transcription factors regulated, so that they are not permanently bound to DNA, continuously promoting gene transcription? We will examine the NF-ĸB transcription factor (remember you learned a little about this protein in the first Jmol tutorial on protein structure) as a case study to address these questions. You should be able to recognize and apply the principles learned from the NF-ĸB system to your understanding of other key cellular functions.
Transcription factors are proteins that bind DNA and help recruit RNA polymerase to the DNA so that transcription can occur. Nuclear Factor-kappaB (NF-ĸB) proteins make up a family of eukaryotic transcription factors that promote the transcription of genes that are involved in many cellular processes, such as immune and inflammatory responses, cell development, cell growth, and apoptosis. NF-ĸB proteins are active in a number of disease states, including HIV-infection, cancer, arthritis, chronic inflammation, asthma, neurodegenerative diseases, and heart disease. NF-ĸB is made up of two protein chains, one p50 subunit and one p65 subunit. NF-ĸB is therefore called a heterodimer because it is composed of two different subunit proteins. Click the first button below to see the NF-ĸB heterodimer then the second button to see the p65 protein of NF-ĸB.
NF-kB heterodimer PDB ID: 1vkxIn this model, NF-ĸB is shown in a backbone format. The DNA binding domain is shown in blue and the dimerization domain is shown in purple. The p65 protein is shown in dark colors and the p50 domain (in the heterodimer model) is shown in light colors. The grey amino acids represent a 'linker' region between the DNA binding domain and the dimerization domain. The N (amino) terminus is the darkest blue and the C (carboxyl) terminus is magenta.
p65 protein of NF-ĸB PDB ID: 1vkxIn this model, the p65 protein of NF-ĸB is shown in a backbone format. Again the DNA binding domain is shown in blue, the dimerization domain is shown in purple, and the 'linker' region between the these domains is shown in grey. The N (amino) terminus is the darkest blue and the C (carboxyl) terminus is magenta.
Based on their primary amino acid sequence, proteins adopt various secondary structures. Recall that secondary structures include α helices, β sheets (β sheets are made of individual β strands), and other structures that we haven't talked about much, such as loops (this helps them out with the answer to the regions shown in grey. Click on the button below to view secondary structure of the p65 subunit of NF-ĸB.
p65 secondary structure PDB ID: 1vkxWhat type of secondary structure are the regions shown in red?
What type of secondary structure are the regions shown in yellow?
What are the regions shown in grey?
What type of bond determines the folding into secondary structures?
Recall that interactions between residues (R groups) are determined by primary and secondary structure, and these residue interactions determine tertiary structure (protein folding) and allow for the formation of functional domains. In the case of NF-ĸB, proper protein folding allows for the formation of the two domains mentioned above: the dimerization domain and the DNA binding domain. Importantly, the proper folding of NF-ĸB also allows it to bind an inhibitor protein called IκBα. Note that the DNA binding and IκBα binding cannot occur until p65 and p50 dimerize (adopt quaternary structure).
As you learned earlier, NF-κB is a heterodimer, meaning it is composed of two distinct monomer or subunit proteins (the p50 and p65 proteins). Residues from each subunit protein bind via hydrogen bonds and van der Waals interactions (Huxford et al 1998). Click on the button below to see the quaternary structure of NF-κB.
NF-kB heterodimer PDB ID: 1vkxAgain, the dimerization domain is shown in purple and the DNA binding domain is shown in blue. The p65 monomer is in dark colors and the p50 monomer is in light colors. The amino acids that interact at the dimer interface are displayed in ball and stick.
Locate two amino acids that you predict interact to help stabilize the dimer (you will need to zoom in).
Quaternary structure (that is, both the p65 and p50 proteins) must be present for NF-ĸB to be functional.
Why do you think NF-ĸB can't function as a monomer?
In its resting (inactivated) state, NF-ĸB is found in the cytoplasm, bound to IκBα (the inhibitor protein, see Fig. 2, step 1). When signals induce degradation of IĸBα, through phosphorylation, ubiquination (Ubn), and proteasomal degradation (Fig. 2, step 2-3), NF-ĸB is then free to translocate to the nucleus, bind DNA, and thus activate gene transcription (Fig. 2, step 4). We will go through these steps in more detail in the following sections.
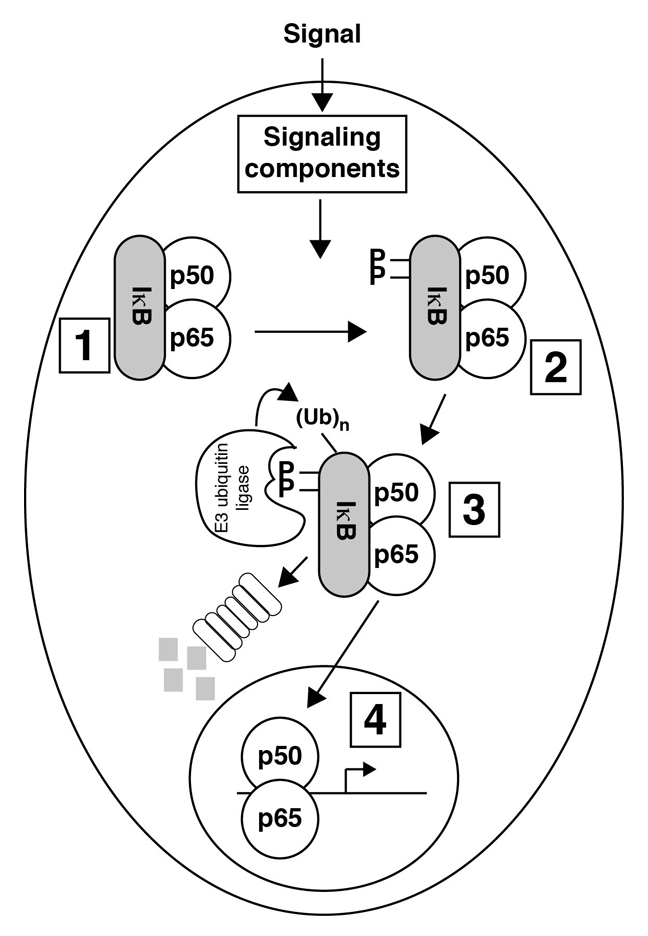
In the schematic diagram shown in Fig. 2, which components represent NF-ĸB?
Both p50 and p65 contain DNA binding domains (regions that interact with DNA). Specific amino acids of the p50 and p65 proteins make interactions with the bases of the nucleotides in the DNA. The following model shows the amino acids that bind DNA in both the p65 and p50 proteins. You will see DNA (white) appear after a few seconds.
NF-ĸB DNA binding amino acids PDB ID: 1vkxIn this model, the DNA binding domains are shown in blue. Four of the amino acids in each subunit protein that bind DNA are shown.
Think about the residues of NF-ĸB that interact with DNA. What properties do you think these residues have?
Do these amino acids bind in the DNA's major or minor groove?
Note how tightly the DNA fits into the space between the two protein chains. The negatively charged DNA has to be very close to the positive residues of NF-ĸB in order for DNA-binding to occur. You can see also why NF-ĸB cannot bind DNA until after it has dimerized.
The p65 protein also contains a transactivation domain that recruits other transcription factors to the DNA to initiate gene transcription. Unfortunately, this region is not part of the crystallized structure.
How is NF-κB transcriptional activity regulated? As you just read, NF-κB is bound to the inhibitor protein, IκBα, in the cytoplasm. Click the button below to see NF-κB bound to IκBα.
IĸBα binding to NF-ĸB PDB ID: 1vkxNotice that in this model, the dimerization domains of both subunit proteins are shown (light and dark purple), but only the DNA binding domain of the p65 subunit is shown (in dark blue). This is because the DNA binding domain of the p50 subunit was not crystallized in this particular structure.
IκBα binds near the NF-κB dimer interface via van der Waals interactions, salt bridges, hydrophobic interactions, electrostatic interactions, and hydrogen bonds. The major structural feature of IκBα is a set of six ankyrin repeats (α helices separated by loops) in the center of the protein. Locate these on your model. When IĸBα is bound, NF-ĸB cannot activate gene transcription for two major reasons:1. The IĸBα protein blocks the nuclear localization sequence (NLS) on NF-κB, so that NF-κB will not localize to the nucleus.2. A negatively charged (highly acidic) portion of IĸBα alters the structure of the DNA binding domains of NF-ĸB so they are no longer able to bind DNA.
Click on the button below to see these features on the model.
IĸBα binding to NF-ĸB prevents DNA binding PDB ID: 1iknThe first two ankyrin repeats of the IĸBα protein block the NLS on NF-κB. In this model, two of the four amino acids in the NLS of p65 are shown in ball and stick (the other two amino acids in the NLS of p65 and the NLS of p50 are not shown because they were not part of the crystallized structure). Locate the two crystallized amino acids of NF-κB on the model shown in the rigth. The side chains of the residues in the NLS interact with IĸBα through salt bridges to acidic groups on IĸBα, thus blocking this signal and preventing translocation of NF-κB to the nucleus. Two of the acidic amino acids of IĸBα are shown in ball and stick- locate these on the model.
When IĸBα binds NF-κB, this alters the structure of the NF-κB DNA binding domains so they are no longer able to bind DNA. The amino acids of the p65 protein that interact with the nucleotides in the DNA are shown in ball and stick on the dark blue portion of the p65 DNA binding domain. Locate these on your model.
One of the regions of IĸBα that binds NF-κB to prevent DNA binding is a negatively charged (highly acidic) portion, why is the charge of this region of IĸBα important?
How might the location and/or activity of NF-κB be affected if you mutate the NLS?
The conformation of the p50/p65 heterodimer is different when bound to IĸBα compared to when it is bound to DNA. Click here: NFkB.html
to compare the two conformations side by side.
How does the conformation of the DNA binding domain change between the two models? Refer to the location of specific amino acids that bind DNA.
When the cell receives a signal, the IĸBα protein is phosphorylated on two serine residues (Ser-32 and Ser-36). This dual phosphorylation recruits the ubiquitination (Ubn) machinery to IĸBα. After IĸBα is ubiquitinated, it is degraded by the proteasome. Degradation of IĸBα exposes the DNA binding domains of p50 and p65 and the NLS of NF-ĸB. The NF-ĸB heterodimer localizes to the nucleus, binds DNA, and activates gene transcription.
What do you think would happen to gene transcription if Ser-32 and Ser-36 were mutated in IĸBα? (assume two missense mutations)
Here is a summary of the regulation of NF-ĸB:1. Cytoplasmic localization of NF-ĸB/IĸBα due to the blocked NLS2. An extracellular signal results in phosphorylation of IĸBα3. Phosphorylated IĸBα is then ubiquitinated (Ubn) and degraded by the proteasome4. The exposed NLS allows NF-ĸB to localize to the nucleus where NF-ĸB binds DNA and activates gene transcription

Putting all of this together, the structure, cellular location (cytoplasm or nucleus), and dynamic interactions of NF-ĸB with other biomolecules, like DNA & IĸBα, all affect gene expression regulation (see Fig. 4).
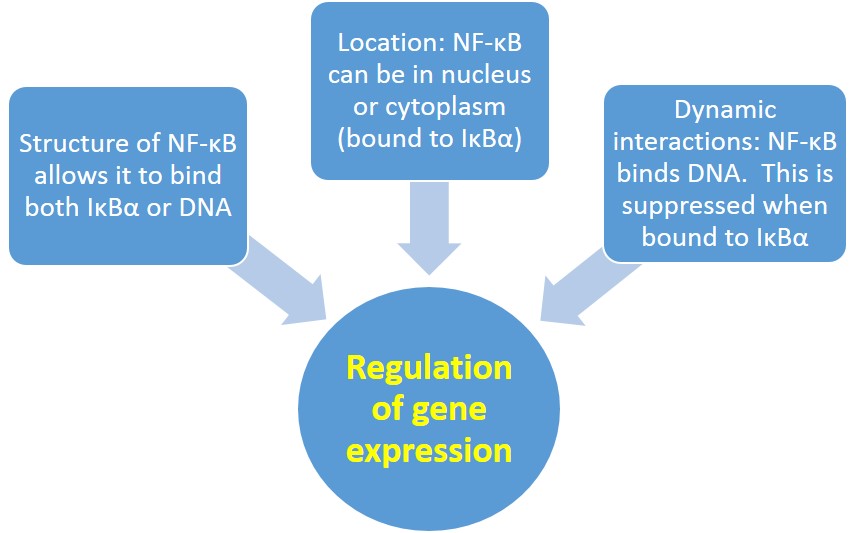
Reflect on how NF-ĸB's structure, cellular location, and dynamic interactions with other biomolecules affect the regulation of gene expression. How does the structure of NF-ĸB allow it to interact with both IĸBα and DNA? How do NF-ĸB's interactions with other biomolecules affect its location in the cell, AND its function as a transcription factor?
Beg AA, Ruben SM, Scheinman RI, Haskill S, Rosen CA, Baldwin AS Jr. (1992) I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa B: a mechanism for cytoplasmic retention. Genes Dev. 6(10):1899-913.
Chen FE, Huang D, Chen Y, and Gosh G (1998) Crystal structure of p50/p65 heterodimer of transcription factor NF-kB bound to DNA. Nature 391: 410-413.
Ganchi PA, Sun SC, Greene WC, Ballard DW (1992) I kappa B/MAD-3 masks the nuclear localization signal of NF-kappa B p65 and requires the transactivation domain to inhibit NF-kappa B p65 DNA binding. Mol Biol Cell. 3(12):1339-52.
Huxford T, Huang DB, Malek S, Ghosh G (1998) The crystal structure of the ikappabalpha/nf-kappab complex reveals mechanisms of nf-kappab inactivation. Cell. 95(6):759.
Jacobs MD & Harrison SC (1998). Structure of an IκBα/NF-κB Complex. Cell. 95(6):749–758.
Lätzer J, Papoian GA, Prentiss MC, Komives EA, Wolynes PG (2007). Induced fit, folding, and recognition of the NF-kappaB-nuclear localization signals by IkappaBalpha and IkappaBbeta. J Mol Biol. 16;367(1):262-74.
Save/Export Your Answers to the Questions in This Jmol Exploration