

This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
Unless otherwise indicated, this section of the Training Guide uses the protein Top 7 based on PDB file 1qys.pdb. Please see Getting Started with Jmol for information on how to download and open PDB files.
Thusfar, every command we have typed into the Command Line has affected the entire protein. However, we can specify parts of a molecule that we want to display differently by using the Select command. Below is a description of commonly used Select commands along with a Jmol button. When using the select command, Jmol will respond with '# atoms selected', where # will be the number of atoms selected. The display will NOT change until you supply additional commands, and those commands will ONLY operate on the parts of the molecule you have selected. To change another part of the molecule, you have to enter a new select command.
Please click the button after each description to see how Top 7 from 1qys.pdb will look with the selected area colored red. (Following the select command, a second command is entered to color the selected portions of the molecule.)
Select all is the default Jmol selection. With select all, every atom contained in a PDB file is selected and will be affected by future commands.
Note: When a select command is executed, Jmol displays the number of atoms selected in the Jmol Console Window. You should get a feel for whether the number of atoms selected is what you expected.
To select every atom, type:
select all
To color this selection red, type:
color red
select all; color redNote: The Jmol buttons of Top 7 shown below have been reset before each set of commands. To do this, use the select all command followed by color gray.
With select backbone, only the backbone atoms will be affected by future commands.
To select the backbone, type:
select backbone
To color this selection dodger blue, type:
color dodgerblue
Note that preset Jmol colors do not have a space between words in the command line.
select backbone; color dodgerblueThe commands above will select all atoms in the peptide backbone - so the N-C-C=O atoms. However, if you are displaying the alpha carbon backbone, in which a line (backbone) connects adjacent alpha carbon atoms (colored cyan in the backbone here: N-C-C=O), you should use the following commands:
select :a
color backbone cyan
With select sidechain, only the atoms that are part of sidechains in a protein will be affected by future commands.
To select the sidechains, type:
select sidechain
To color this selection lemon chiffon, type:
color lemonchiffon
select sidechain; color lemonchiffonWith select hydrophobic, only the hydrophobic amino acids in a protein will be affected by future commands.
To select hydrophobic, type:
select hydrophobic
To color this selection yellow, type:
color yellow
select hydrophobic; color yellowWith select helix, all atoms that are part of alpha helices will be affected by future commands.
To select alpha helices, type:
select helix
To color this selection red, type:
color red
select helix; color redNote: Jmol does not recognize 'helices' (plural) but only 'helix' (singular).
With select sheet, all atoms that are part of beta pleated sheets will be affected by future commands.
To select beta pleated sheets, type:
select sheet
To color this selection yellow, type:
color yellow
select sheet; color yellowWith select nucleic, all atoms that are part of nucleic acids (DNA, RNA) will be affected by future commands.
To select nucleic acids, type:
select nucleic
To color this selection salmon, type:
color salmon
Note: The Top 7 protein does not contain any nucleic acids and therefore nothing is colored when the select nucleic command is used. A zinc finger protein based on the file 1zaa.pdb does contain nucleic acids and therefore will have areas colored red. Click the button below to see select nucleic on a zinc finger protein.
select nucleic; color salmon - Zif268 PDB ID: 1zaaWith select water, all water molecules will be affected by future commands.
To select water molecules, type:
select water
To display water molecules, type:
spacefill 1.2
To color this selection red, type:
color red
select water; spacefill 1.2; color redNote: Hydrogen atoms are not displayed in pdb files that are obtained via X-ray crystallography. Therefore water molecules appear as a single oxygen atom. Since the oxygen is not visually bonded to another atom, the only way water molecules can be displayed is in spacefill format.
With select :, followed by the letter of the chain you would like to select, only the chain you select will be affected by future commands.
To select chain A, type:
select :a
To color this selection green, type:
coor green
Note: The Top 7 protein is a protein with only a single chain, so all atoms are colored red with the commands used above. Hemoglobin based on the file 1a3n.pdb contains four chains so the commands above will only color a portion of the structure. Click the button below to see select :a on hemoglobin.
select :a; color green - hemoglobin PDB ID: 1a3nWith the Residue Number select command, you can select a specific amino acid or a range of amino acids that all future commands will affect.
To select residue number 58, type:
select 58
To color this selection purple, type:
color purple
select 58; color purpleNote: You can also select an amino acid by its three letter abbreviation followed by the residue number. For example, instead of select 58 as shown above, you could also use select lys58 because the amino acid at residue number 58 in the Top 7 protein is lysine. There are two advantages to including the residue name. First, if you accidentally mistype the residue number, if there is a mismatch between the residue name and number, Jmol will return '0 atoms selected'. Second, if you keep a record of your work, including the residue name may be helpful in remembering why you displayed the residue. For more information on documenting your model design, see the tutorial Maintaining a Record of Your Work.
To select residues 20 through 30, type:
select 20-30
To color this selection orange, type:
color orange
select 20-30; color orangeWith the Atom Number select command, you can select a specific atom number or range of atoms that all future commands will affect.
To select atom number 317, type:
select atomno=317
or you may use the shortcut:
select @317
To color this selection magenta, type:
color magenta
select @317; color magentaWith an alteration to the Atom Number select command, you can select a range of atoms that all future commands will affect.
To select atom number 303 through 410, type:
select atomno>=303 and atomno<=410
Note that the shortcut @ does NOT work with this selection!
To color this selection lawn green, type:
color lawngreen
select atomno>=303 and atomno<=410; color lawngreenNote: If you do not know the Residue Number, Chain, Amino Acid Type, or the Atom Number you want to select, you can click on the residue in the Display Window. Jmol will provide information on the Console Window regarding the atom you clicked. The image below decodes this information in detail:
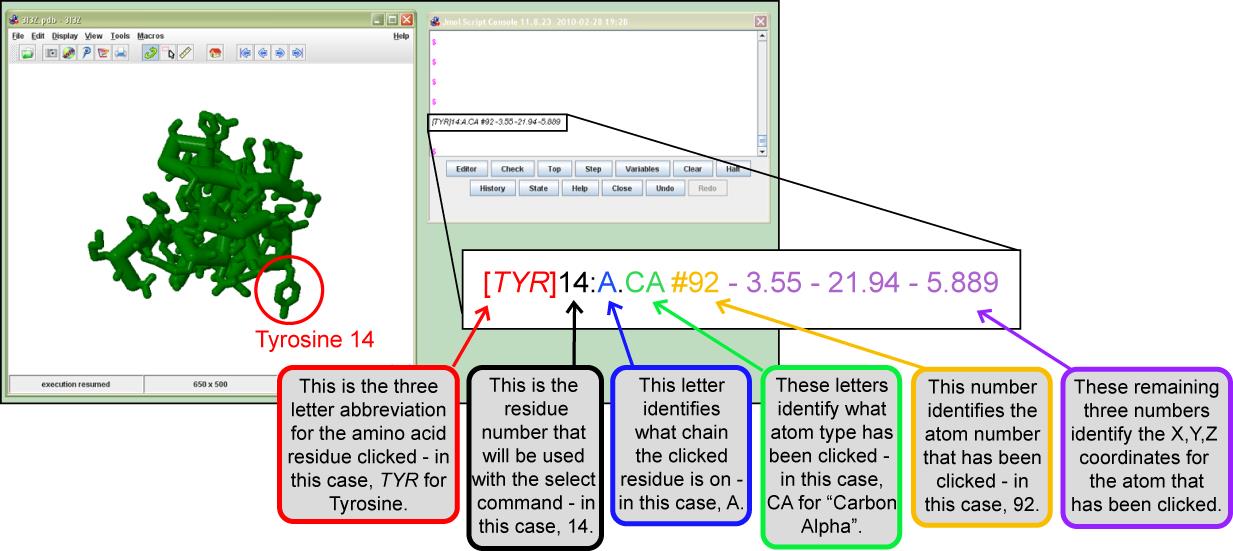
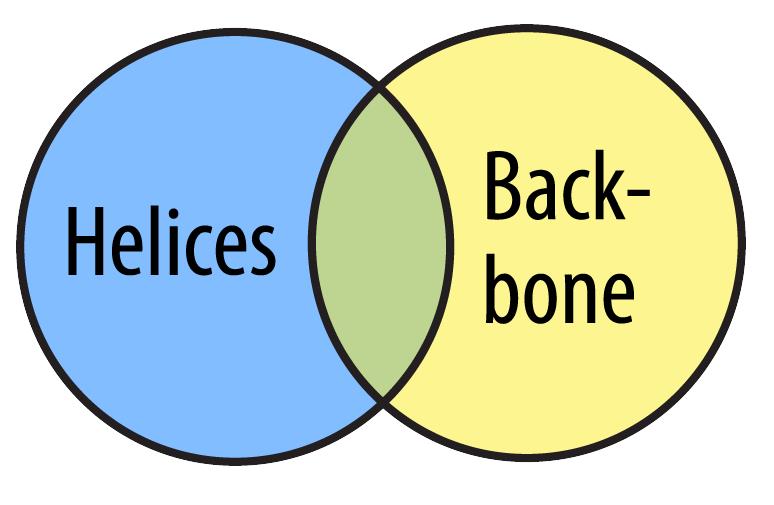
Once you are comfortable using the Select command, you can begin to link together selections by using Boolean Operators. Imagine you are working with two predefined sets, for example, helix and backbone which are illustrated in the Venn diagram below.
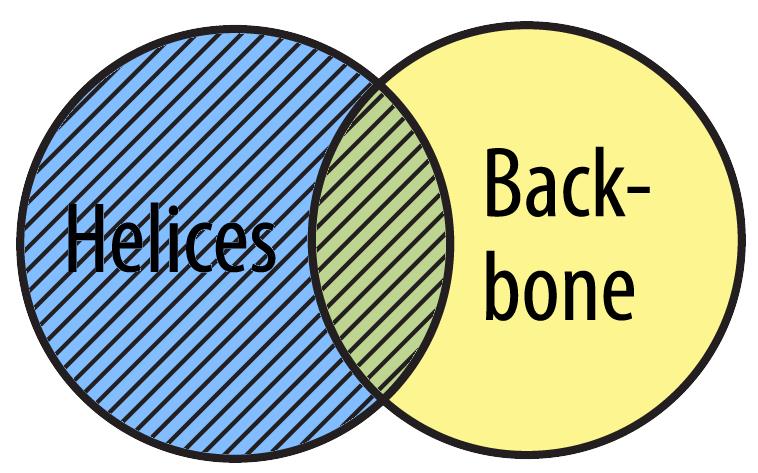
You can select just the helices. This set includes all the alpha helices (including any backbone atoms that happen to be in alpha helices). The area with the hatched lines represents what is selected.
select helix
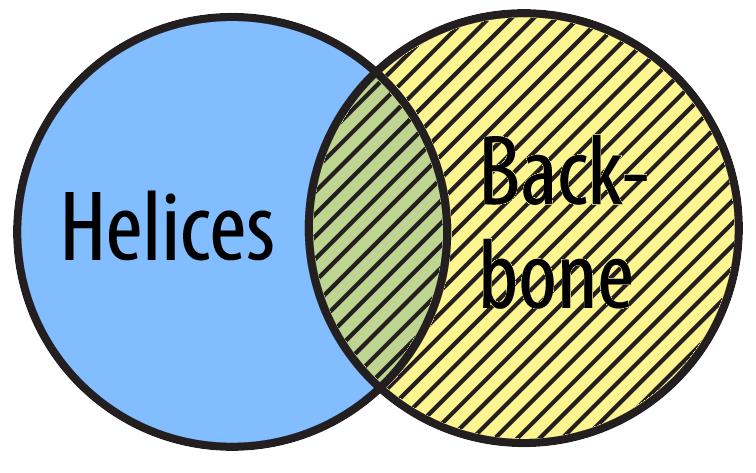
You can also select backbone. This set includes all the atoms that are part of the backbone (including any alpha helix atom that also happen to be backbone atoms).
select backbone
You can use the or Boolean Operator to select all the amino acids that are either part of alpha helices or the atoms that are part of the backbone.
select helix or backbone
The phrase 'select helix or backbone' means 'select all atoms that are either a helix or in the backbone (or both)'.
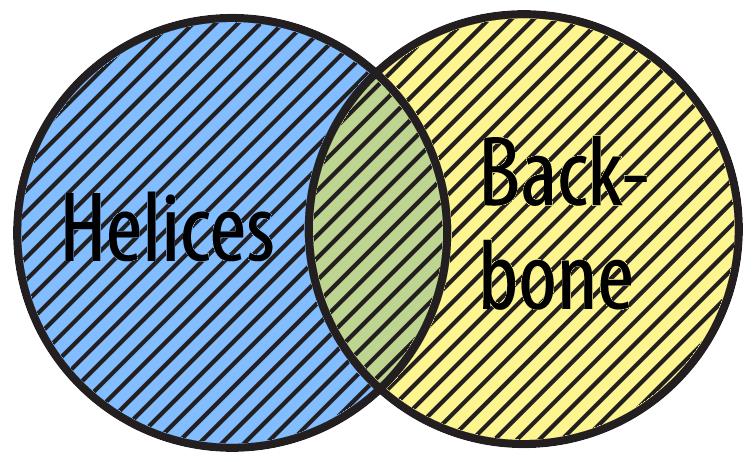
Jmol shortcut: Instead of using the operator or, you may also separate items with a comma:
select helix, backbone
is equivalent to:
select helix or backbone
You can use the and Boolean Operator to select all the amino acids that are both backbone amino acids as well as amino acids in alpha helices.
select helix and backbone
The phrase 'select helix and backbone' means 'select all atoms that are both part of a helix AND part of the backbone'.
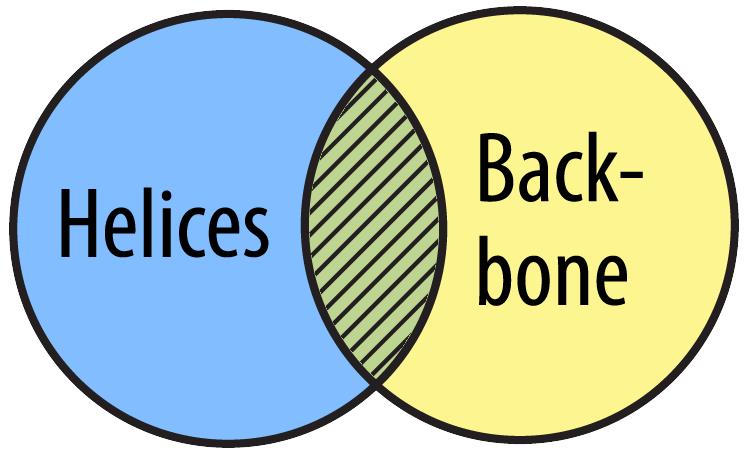
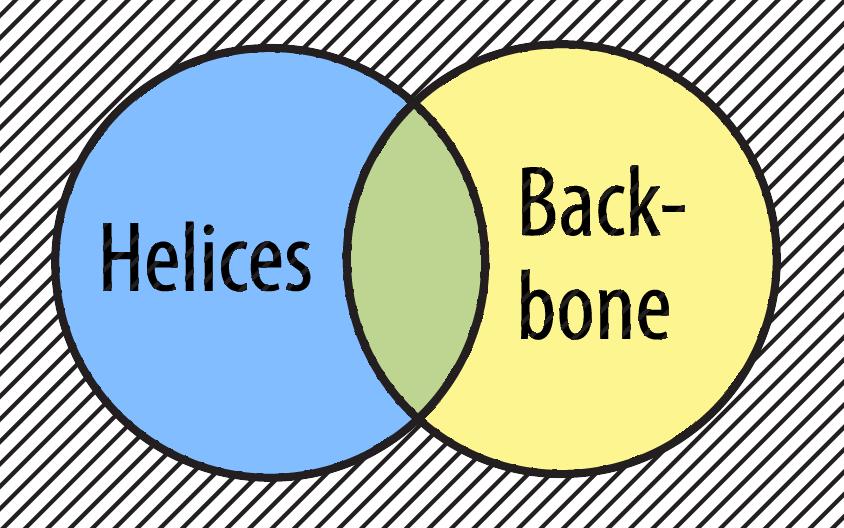
You can use the not Boolean Operator to select only the atoms that are not part of helices and not part of the backbone.
select not helix and not backbone /span>
The phrase 'select not helices and not backbone' means 'select all atoms that are neither a helix nor part of the backbone'.
When using select and Boolean operators in Jmol, occasionally '0 atoms selected' will be displayed in the Console Window. This means the selection is invalid. Consider for instance, the command:
select helix and sheet
The phrase 'select helix and sheet' means 'select all the atoms that are BOTH helices AND sheets'. In the diagram below, you'll see that these are two mutually exclusive sets of atoms. There are no atoms that fulfill this selection, and Jmol will diplay '0 atoms selected'.
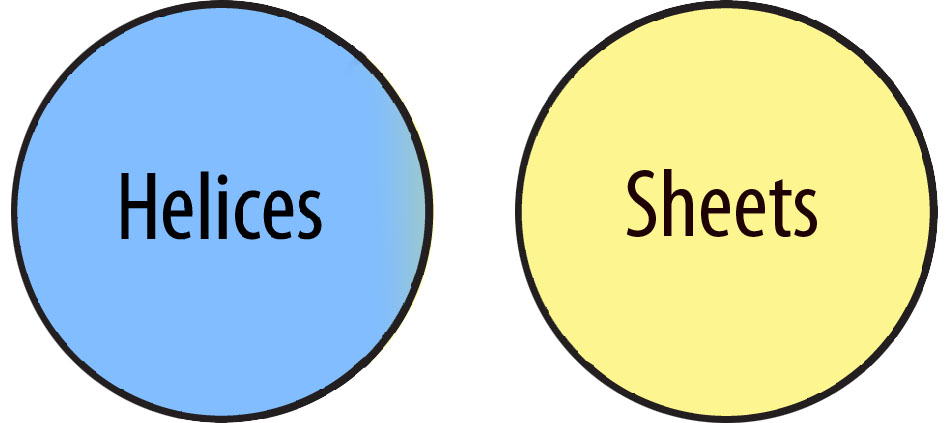
Parentheses can be used in a select command when combining operators to insure that the proper selection occurs. Here are some sample commands that are frequently used in designing models:
select :b and (his63 or his92) and (sidechain or alpha)
This selects the sidechain and alpha carbon atoms in two residues, his63 and his92, on chain b of the protein.
select (:a or :c) and sheet
This command selects the sheets in chains a and c of the molecule.