

This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
The interaction between the substrate and enzyme is highly specific. Even a slight change in shape of either the substrate or the enzyme may alter the efficient and selective ability of the enzyme to catalyze the reaction.
Human cells have more than 100,000 different proteins, many of which are enzymes. There are between five million and two trillion molecules in a typical human cell. Each enzyme will catalyze a reaction with only a small subset of all these molecules in the cell. How does the enzyme know with which molecules (substrates) to react?
To explore this question, you will use a schematic model of an enzyme and substrate (Substrate Specificity Kit). If you do not have access to the kit, images and videos in this exploration will guide you.
Throughout the activity, you will see a question mark icon. This indicates that a question needs to be answered. You can either type your response directly into the exploration (there's a button at the end to export your answers), or you can download and print the worksheet.
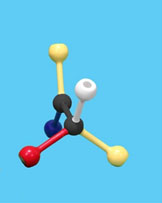
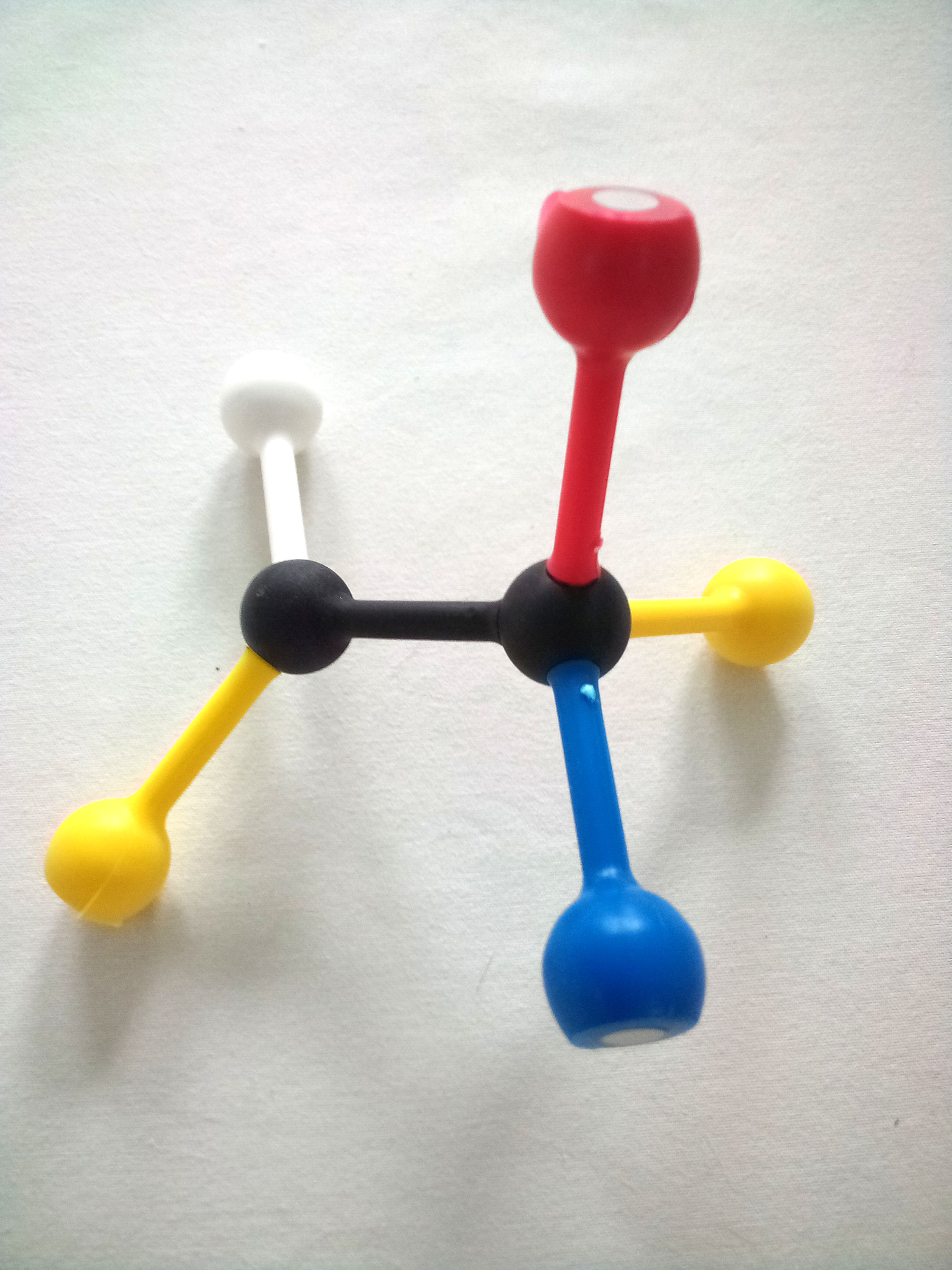
Use the images and instructions below to assemble the substrate.


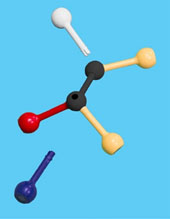
The structure you have assembled is a generic substrate. The colored pieces represent properties of various functional groups or atoms.
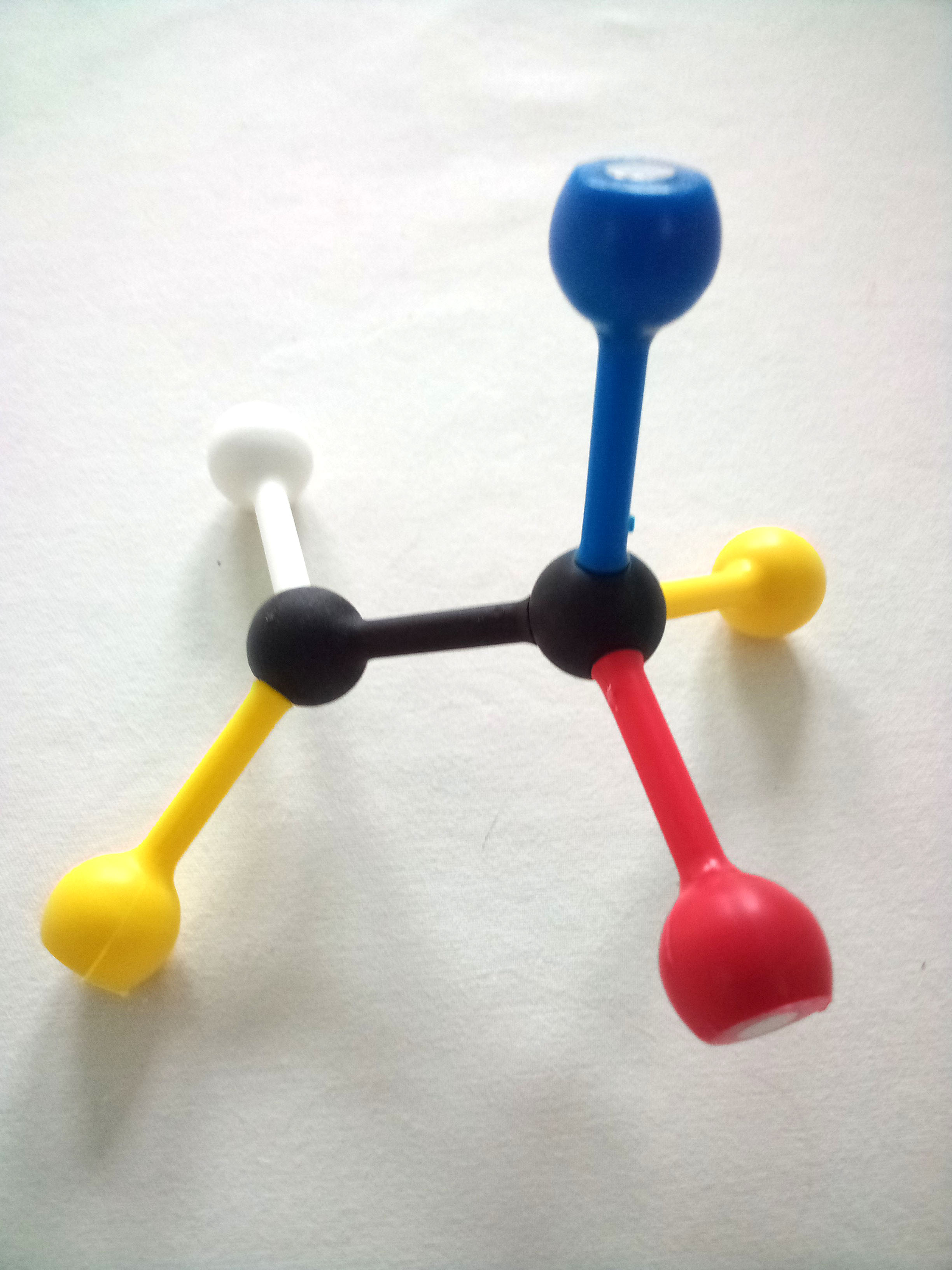
1. Draw and label your substrate with the appropriate colors/chemical properties.
Use the images and instructions below as a guide to assemble the enzyme.


2. What does the toober represent?
3. What do the clips on the toober represent?
The colored clips represent specific amino acid side chains (R groups) that are in the enzyme and will make up the active site of the enzyme. As in the substrate, each color represents a specific chemical property:
4.For each clip, choose a specific amino acid that has the same chemical properties as the clip. List the color of the clip and the amino acid that clip represents for each of the clips on your protein.
Now you will make the enzyme-substrate complex by folding the toober around the substrate. Make sure to match up the metal clips and the groups on the substrate correctly.
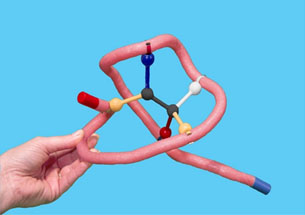
5. What color ball (functional group) should match with the yellow clip?
6. What color ball (functional group) should match with the white clip?
7. What color ball (functional group) should match with the red clip?
8. What color ball (functional group) should match with the blue clip?
In the past, it was thought that both enzyme and substrate were rigid structures with the substrate (the key) fitting perfectly into the active site (the lock). The lock and key model implies that the enzyme has an optimum substrate, whereas all other substrates fit less perfectly. We now know that some enzymes can catalyze a reaction on a range of different substrates. The induced fit model proposes that the substrate induces the active site to take on an ideal shape to accommodate it. This new model explains why some enzymes can catalyze a wide range of substrates. Additionally, the substrate is not a passive structure in the process. Bond rotation may occur to fix the substrate in a particular conformation allowing for catalysis.
Gently remove the substrate from the enzyme. Rotate the functional groups around the movable bond between the two black spheres and try to dock the new configuration back into the enzyme.
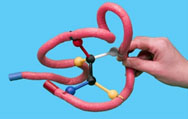


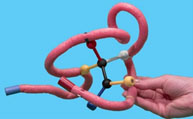
9. Describe how well the substrate binds in the active site after rotating the bond.
10. What is the purpose of holding the substrate in a specific conformation?
Return the substrate to the original configuration.
Imagine that a mutation has occurred that caused a variant form of the enzyme. To simulate a mutation, remove one of the clips from the active site.
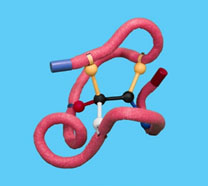
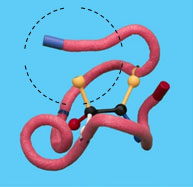
11. What is the effect of the loss of one of the active site side chains on the strength of the binding of the enzyme to the substrate?
Return the enzyme to the original conformation.
Stereochemical specificity means that an enzyme can only catalyze one enantiomer. To determine if your enzyme has stereochemical specificity, you will make the enantiomer and determine if it fits into the active site or not.
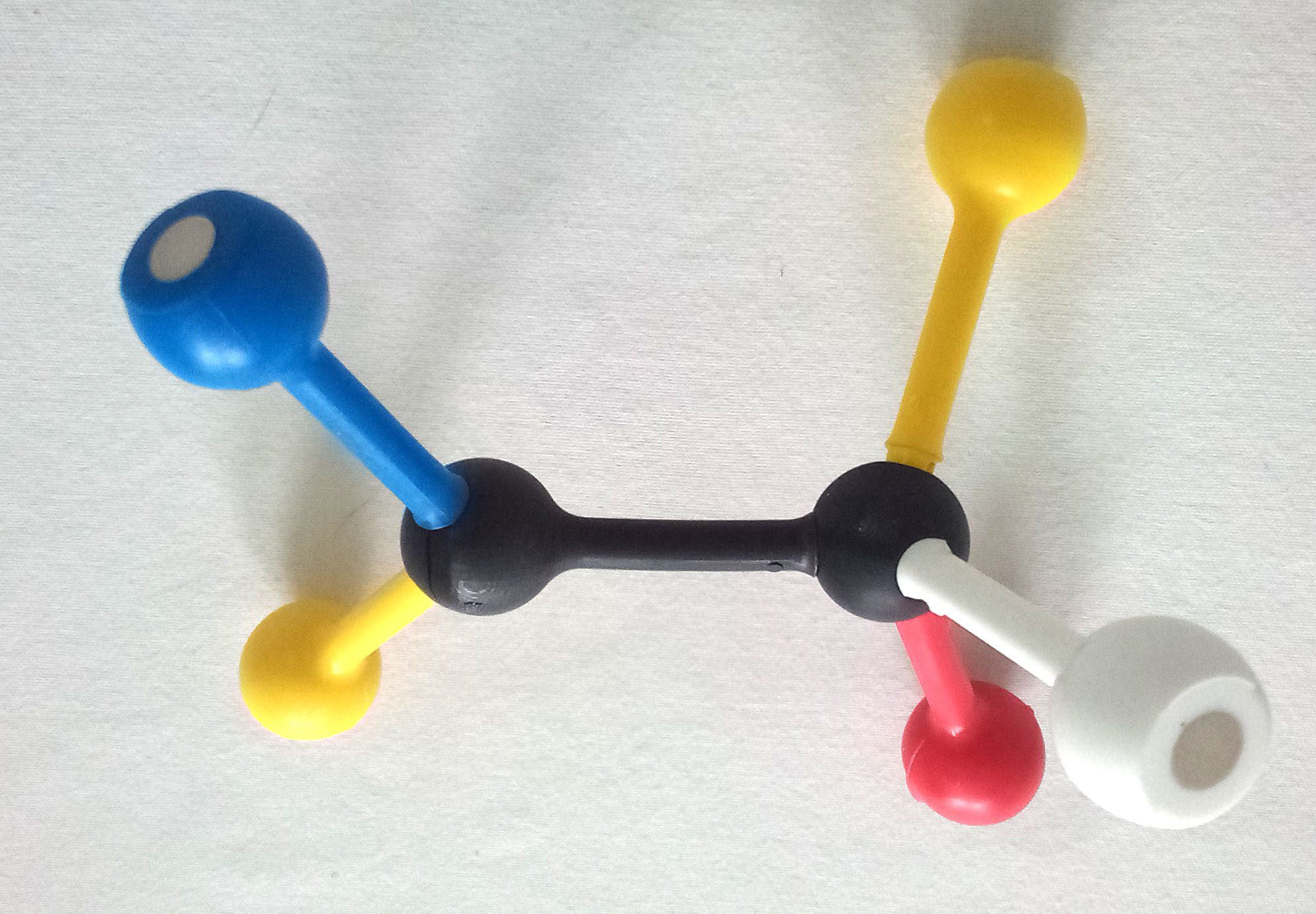
Gently remove the substrate, WITHOUT disturbing the enzyme. Then exchange two of the groups on the 4-hole sphere to make the mirror image of the original substrate – the enantiomer (mirror image).
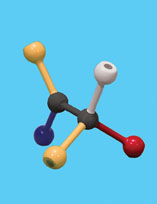



Next, try to fit the enantiomer into the active site. Do not adjust the active site!
12. Draw the enantiomer that you made.
13. Draw the enantiomer bound to the enzyme.
14. Was your enzyme stereospecific or not? Explain.
Return your substrate to its original form.
Absolute specificity means that an enzyme can only catalyze one and only one substrate. To determine if your enzyme has absolute specificity, trade your substrate with someone else, making sure that theirs is NOT either the same as your or an enantiomer of yours.

15. Draw your substrate and the one that you borrowed.
Try to fit the borrowed substrate in your active site.

16. Draw the borrowed substrate in your active site.
17. Describe the results.
The following two buttons explore the active site of two enzymes. In both structures, the protein backbone is in pale yellow, sidechains in the active site are in soft cpk (light gray carbon, pink oxygen and blue nitrogen) and the substrate is in cpk (gray carbon, red oxygen and blue carbon). Use a 'click and drag' to rotate the structures to see the interactions between the enzyme and substrate. You can also zoom in and out using shift-left mouse up and down.
Glutamate Dehydrogenase PDB ID: 1bgv18. What types of interactions do you see between glutamate dehydrogenase and its substrate glutamate?
19. What types of interactions do you see between hexokinase and its substrate glucose?
George E. P. Box, a British mathematician and a professor of statistics at the University of Wisconsin, has said, 'All models are wrong; some models are useful.' While models help us to build our conceptual understanding of a topic, they are a simplification of reality. Answer the following questions to evaluate the model you have just used.
20. What new information have you discovered by using this model?
21. What have you learned or clarified with this activity?