

This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
In this activity you will learn how DNA is used as a template to code for a protein, and how that protein is synthesized. If you have access to the models, you will work with them to complete the activity. Otherwise, the images and videos included below will guide you through the activity. If you are working with models, your instructor will indicate whether you will work alone or in groups.
This icon indicates you are to provide a response. You may enter your response in the activity, then download a file with your answers at the end of the tutorial. Alternately, you may download and print a paper copy of the worksheet. Question numbers are included to allow you to easily transition from the tutorial to the worksheet.
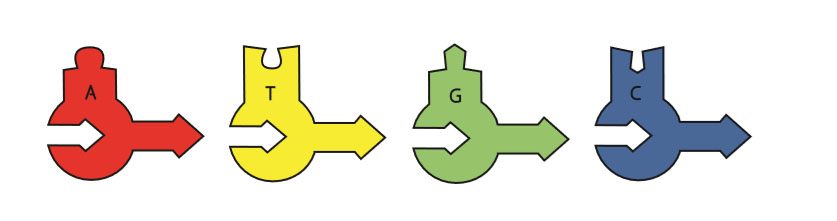
You will be using schematic models in this activity. Deoxynucleotides (A, C, G and T found in DNA) have a rounded base, and ribonucleotides (A, C, G and U found in RNA) have a square base:
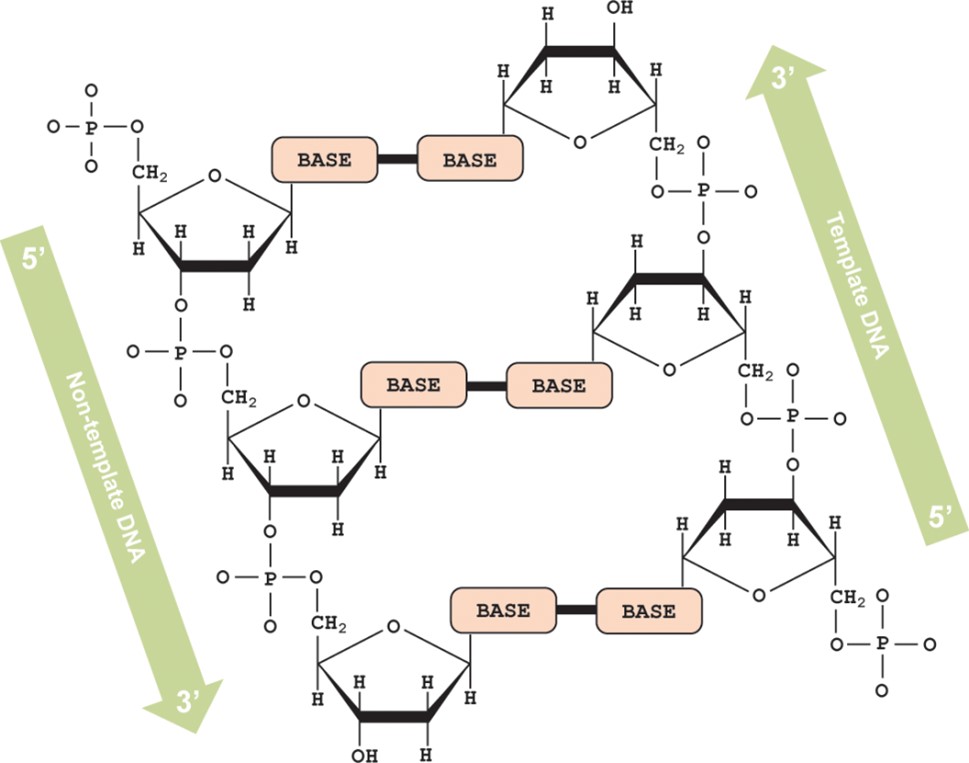
As a reminder, the schematic model you are using is a representation of DNA. Below is a depiction of the structure of DNA, showing that the two strands run in opposite (antiparallel) directions. In the schematic model, the arrow on the base represents the 3' end of the molecule.
Using the rounded DNA foam pieces and following the code listed below, create a non-template strand of DNA (beginning with CC on the 5' end – the lower strand of sequence 1). On the DNA backbone the sugar end is the 3' end (arrow end of the foam piece) and the phosphate end is the 5' end (hole to which arrow attaches). In order for DNA to be interpreted correctly the 5' → 3' direction must be maintained. Refer to the figure above to ensure correct initiation of the protein synthesis process. Recall the antiparallel nature of the DNA molecule.
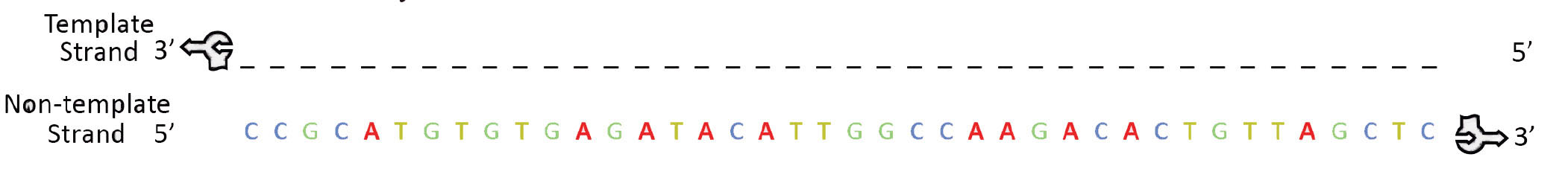
Build the template strand of DNA to create a double stranded DNA model. Attach the two strands together following the rules of complementary DNA base pairing.
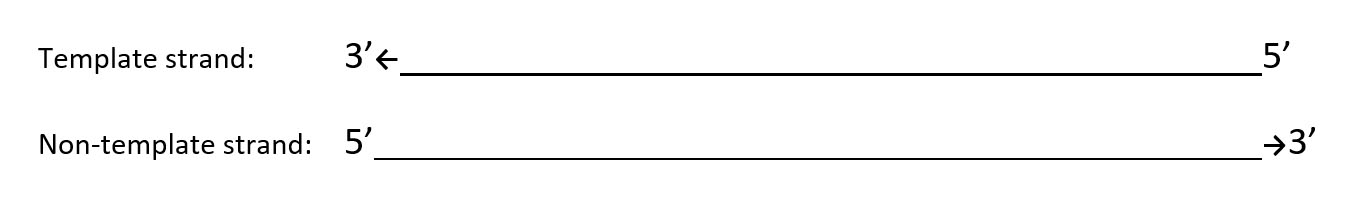
1. Fill in the correct base pairs in the non-template and template strands below.
2. Recalling from the lesson on DNA structure identify the type of bond that holds the two strands of DNA together.
Compare and contrast the foam model to the DNA Discovery Kit model or DNA Starter kit model on display. If you don't have access to the models, look at the images below.
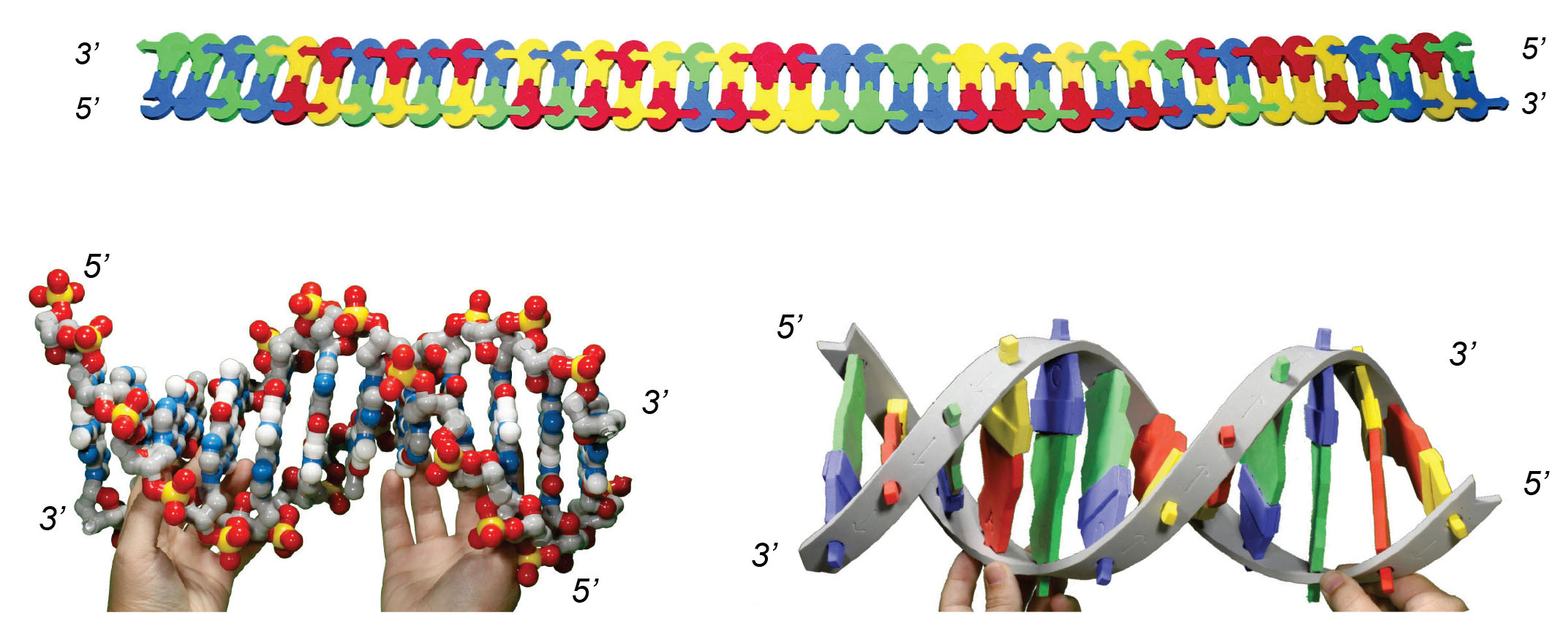
3. Identify two similarities and two differences between these models.
RNA polymerase assembles the mRNA only in its 5' → 3' direction. In order for this to properly occur, the template strand of DNA must be oriented in the top slot with the 3' end (arrow end) entering the polymerase first. (Please refer to the photo to ensure proper setup.)
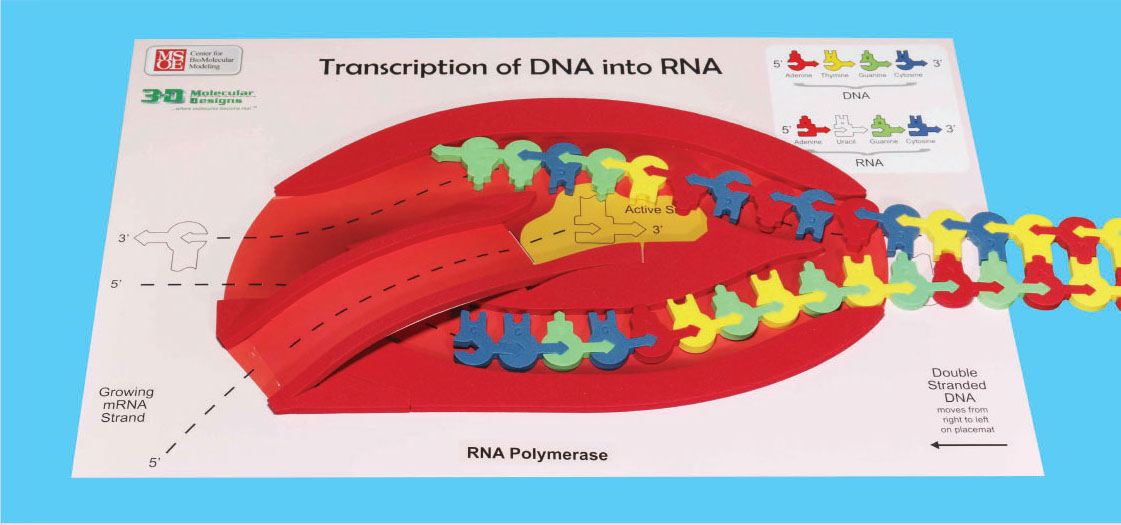
The template strand is the strand of DNA that RNA polymerases 'reads' to build an RNA strand. The nontemplate strand is the other strand of DNA.
Gently feed the DNA into the RNA polymerase from the right, making sure the bottom left of the DNA starts with the sequence CC. u>Check the markings on the Transcription placemat to make sure the 3' and 5' ends of the DNA are in the correct orientation! As you feed the DNA into the RNA polymerase, a foam bumper simulates the breaking the hydrogen bonds between the base pairs (refer to photo), separating the two strands of DNA into separate channels. When DNA opens up, the RNA polymerase uses the template strand of DNA to synthesize the mRNA.
4. Label the DNA template strand and non-template strand in the image below.
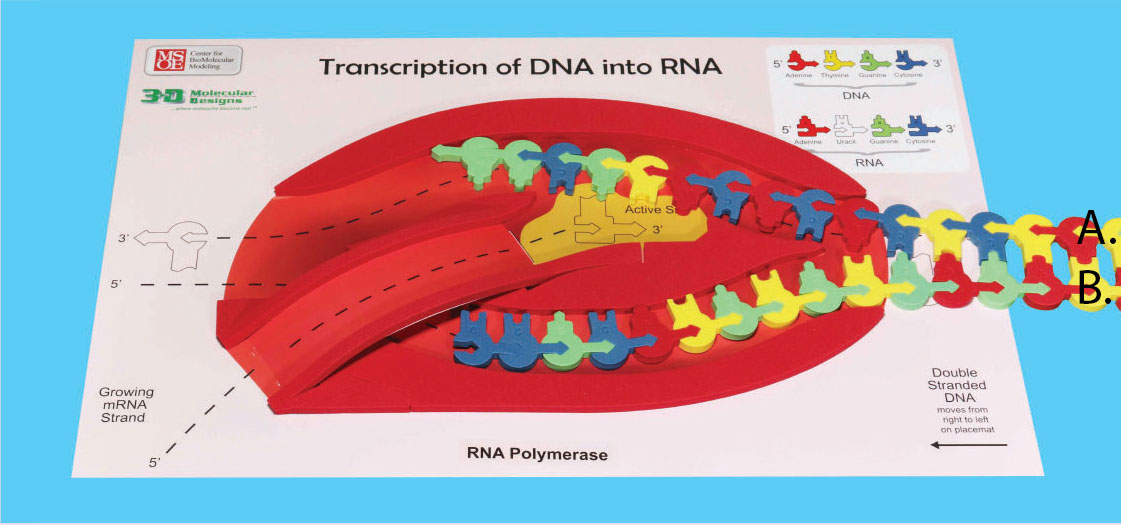
Continue moving the DNA through the RNA polymerase, building your RNA model. As the two DNA strands exit on the left, pinch them together to form double-stranded DNA.
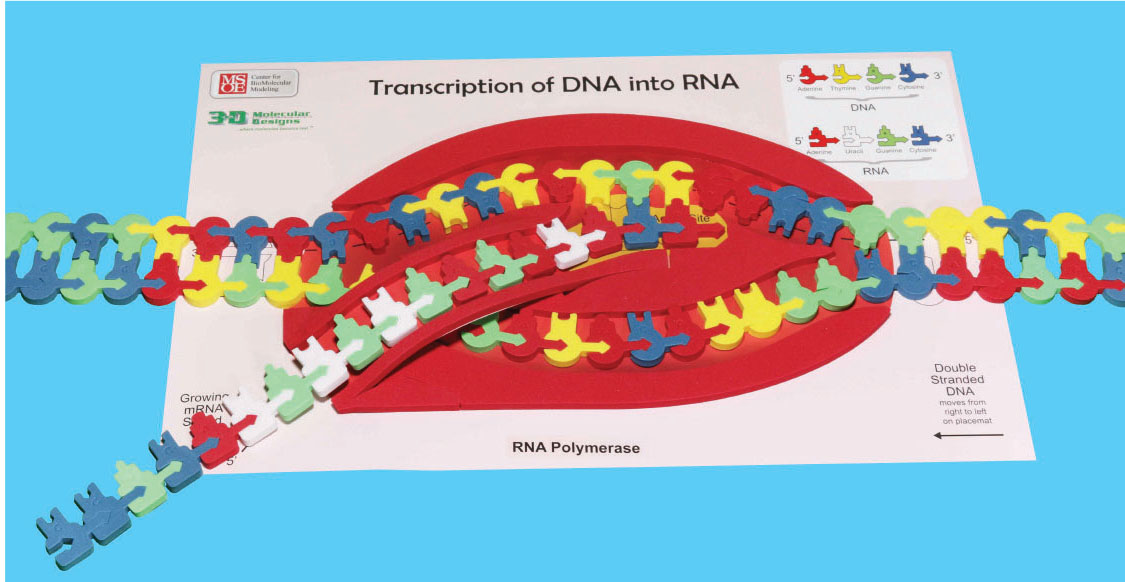
At this point the mRNA will separate from the DNA and may be processed into its final form. The template strand of DNA will rejoin with the nontemplate strand. Complete this step with your model.
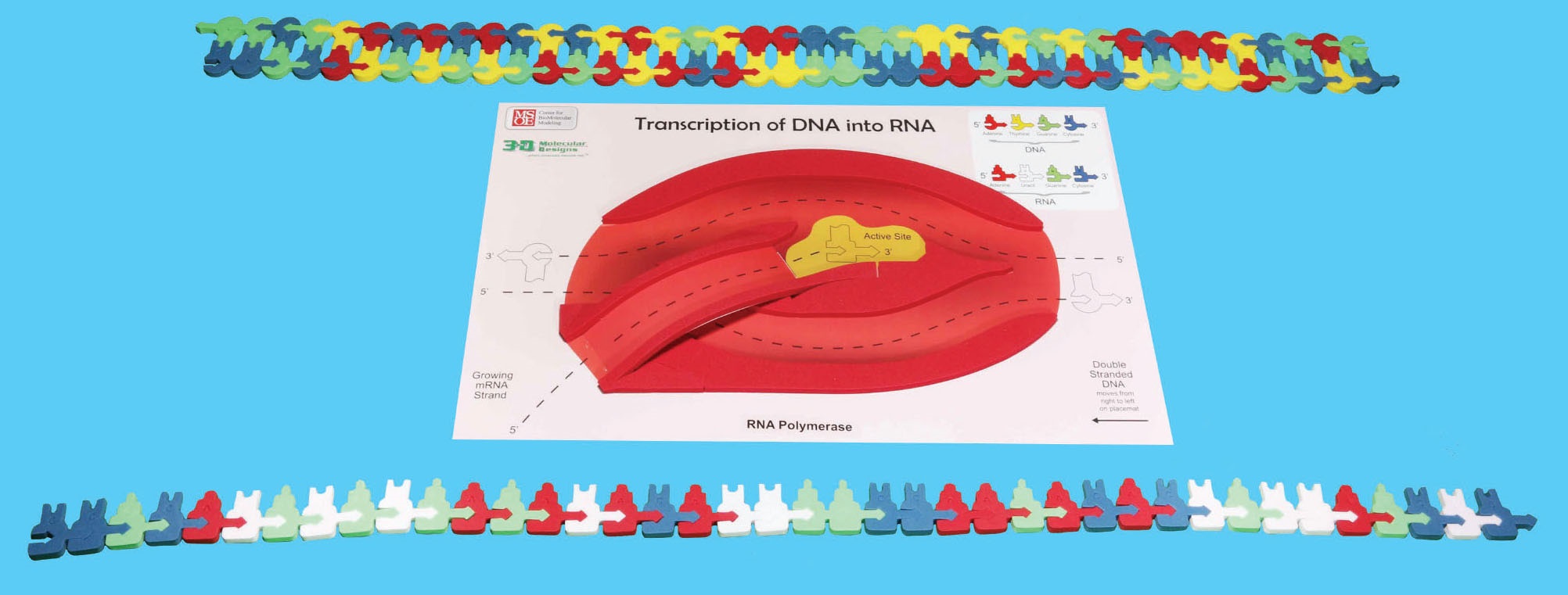
5. Using the mRNA model you just made record the correct sequence of mRNA base pairs:
6. What type of intermolecular force (NOT bond) is broken when mRNA separates from DNA and what characteristic of this allows for this separation?

In eukaryotic cells the mRNA leaves the nucleus through nuclear pores after being processed into its final form.
STOP: Double check the sequence of the mRNA strand you made. It should match the mRNA on the strip of paper that your instructor has.
For a quick review of transcription, view this stop motion video:
Translation occurs in the cytoplasm of the cell and is defined as the synthesis of a protein (polypeptide) using information encoded in an mRNA molecule. Translation may also be thought of in three stages: (1) initiation, (2) elongation and (3) termination. Messenger RNA (mRNA) has the information for arranging the amino acids in the correct order to make a functional protein.
7. What part of mRNA contains the information to make a protein?
Interpretation of the nitrogen bases in mRNA occurs in groups of threes called a codon. The three bases in one codon will indicate a specific amino acid. The order in which the amino acids are put together depends on the sequence of bases in the mRNA. Typically one mRNA strand will result in a protein (polypeptide strand) that can be hundreds to thousands of amino acids long.
The identity of the amino acids in the protein sequence can be determined using the mRNA strand you created above. In the part of the mRNA that encodes a protein, every three bases (in the 5' to 3' direction) determines a particular amino acid.
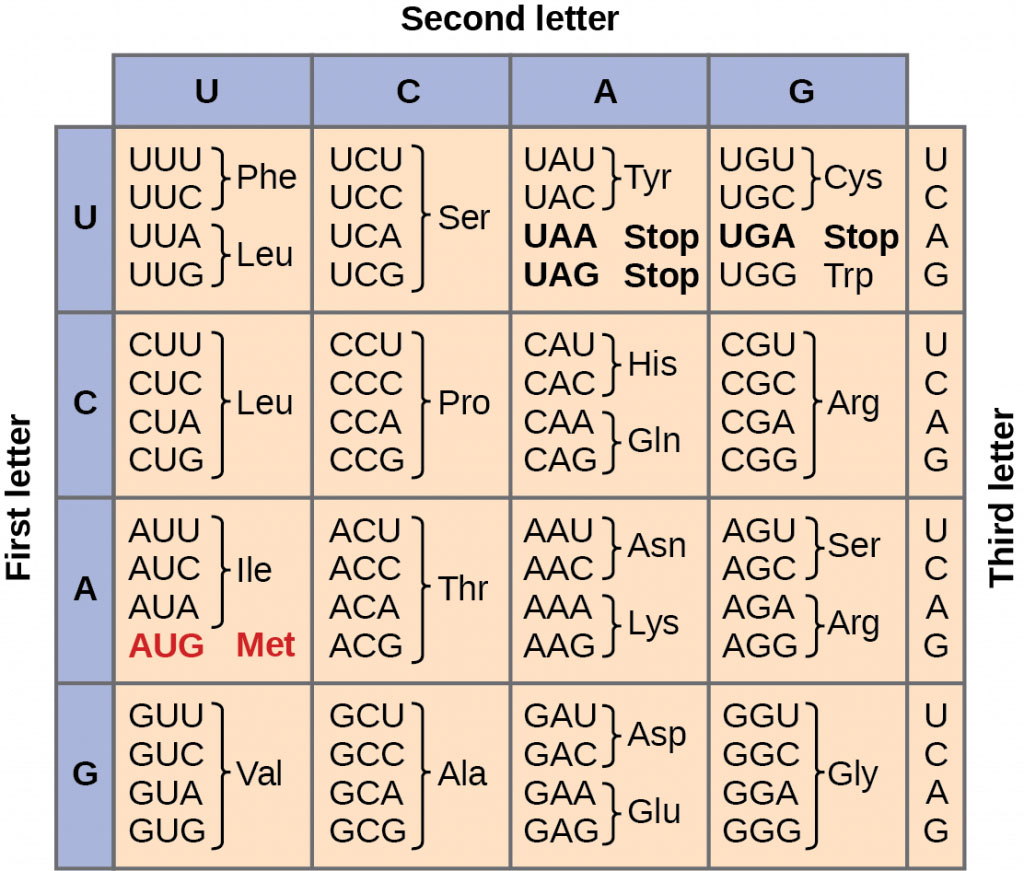
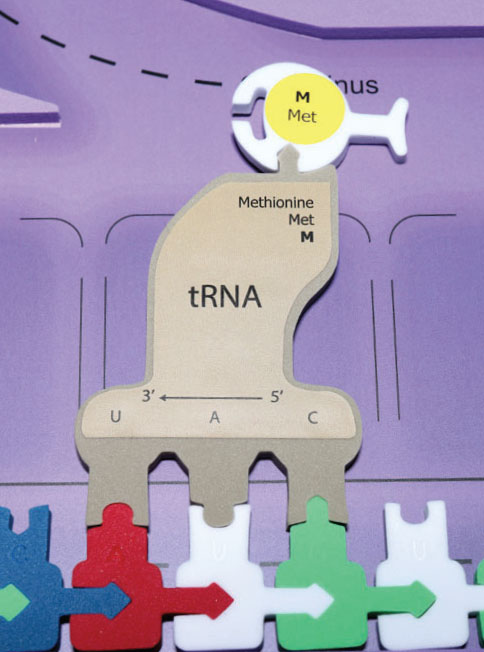
The initiation stage of translation brings together mRNA, a second type of RNA called transfer RNA (tRNA) and the two subunits of a ribosome. Initiation begins with the codon AUG.
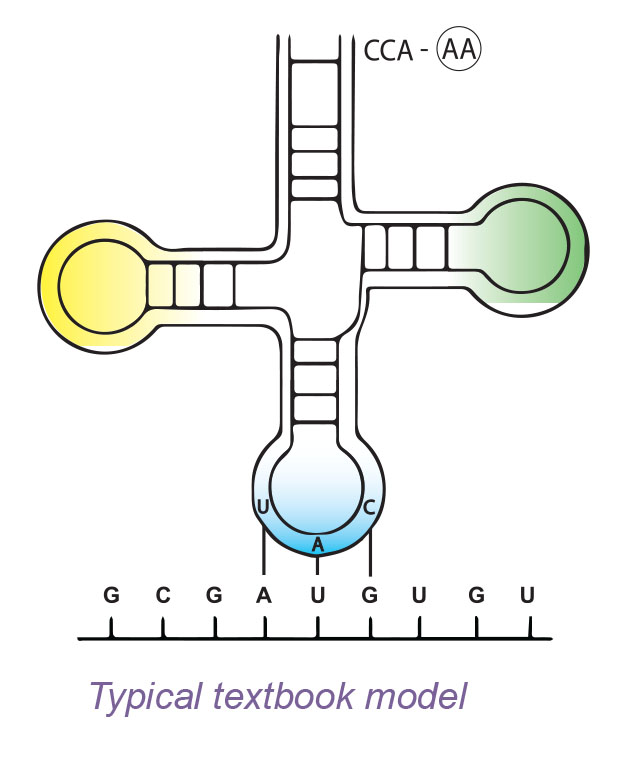
Two functional portions of the tRNA are necessary for protein synthesis to continue. One functional part of tRNA is a series of three nitrogen bases referred to as an anticodon. This anticodon complementary base pairs with the codon of the mRNA. The other functional part of tRNA attaches to a specific amino acid.
Below are crystal structures of phenylalanine tRNA in backbone and spacefill. The blue end of the molecule is where phenylalanine is attached. The green portion of the molecule is the anticodon. In the spacefill model, a few pieces appear to be 'floating'. When the X-ray crystal data was collected, some of the atoms in each molecule were in slightly different locations, so their actual location could not be determined.
Phenylalanine tRNA molecule backbone PDB ID: 4tnaInteresting Note: If one tRNA anticodon variety existed for each mRNA codon specifying an amino acid, there would be 61 tRNAs. In fact, there are only about 45, implying that some tRNAs must be able to bind to more than one codon. Such flexibility is possible because the rules for base pairing between the third nucleotide base of the mRNA codon and the corresponding tRNA anticodon are relaxed. Flexible base pairing at this codon position is referred to as wobble. For example, a tRNA with the anticodon 3'-CGU-5' can base pair with either the mRNA codon 5'-GCA-3' or 5'-GCG-3' both of which code for alanine.
8. Translation begins at a specific codon on the mRNA. What is the three base sequence of the initiation codon?
9. What amino acid is associated with the tRNA that will bind to the mRNA codon AUG?
10. What would be the anticodon for the mRNA codon AUG? Label the 5' and 3' ends of the anticodon.
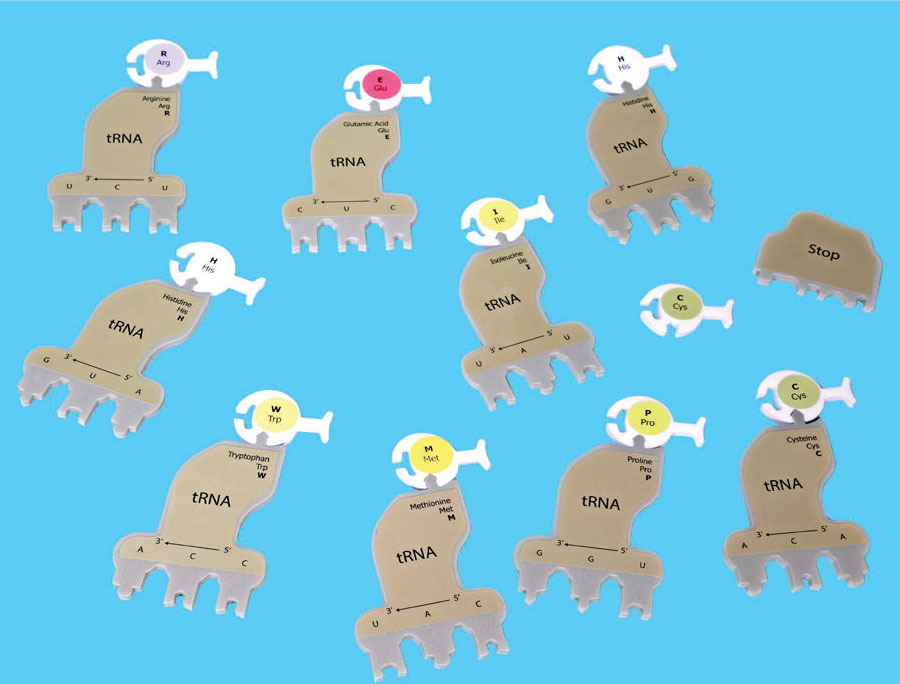
Bond the appropriate amino acids to each of the brown tRNAs. The appropriate amino acid is listed on its corresponding tRNA. The amino acids have different colors which represent their various chemical properties such as acidic, basic, hydrophobic, and hydrophilic. A tRNA molecule with an amino acid attached is called a charged tRNA.
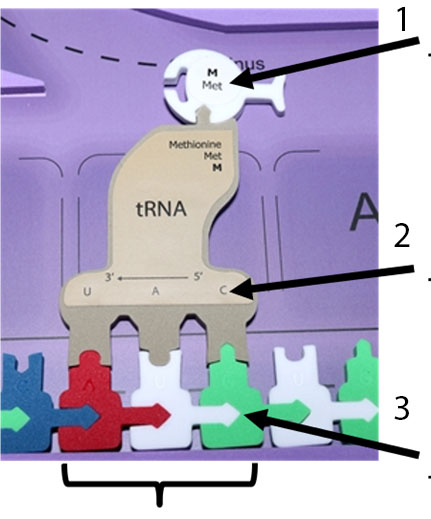
11. Label the codon, anticodon and the amino acid on the figure below.
While the charged tRNA molecules are assembling in the cytoplasm, mRNA moves towards the ribosome. Ribosomal subunits are made in the nucleolus of eukaryotic cells. The resulting ribosomal subunits are exported via nuclear pores to the cytoplasm. Approximately one third of the mass of a ribosome is made up of protein while the rest is composed of a third type of RNA, ribosomal ribonucleic acid (rRNA).
The ribosome consists of two separate parts; the large and small subunits which are unattached when not in use. First, the small ribosome subunit binds to both mRNA and a specific initiator tRNA bearing the amino acid methionine. The attachment of the large ribosomal subunit completes the translation initiation complex.
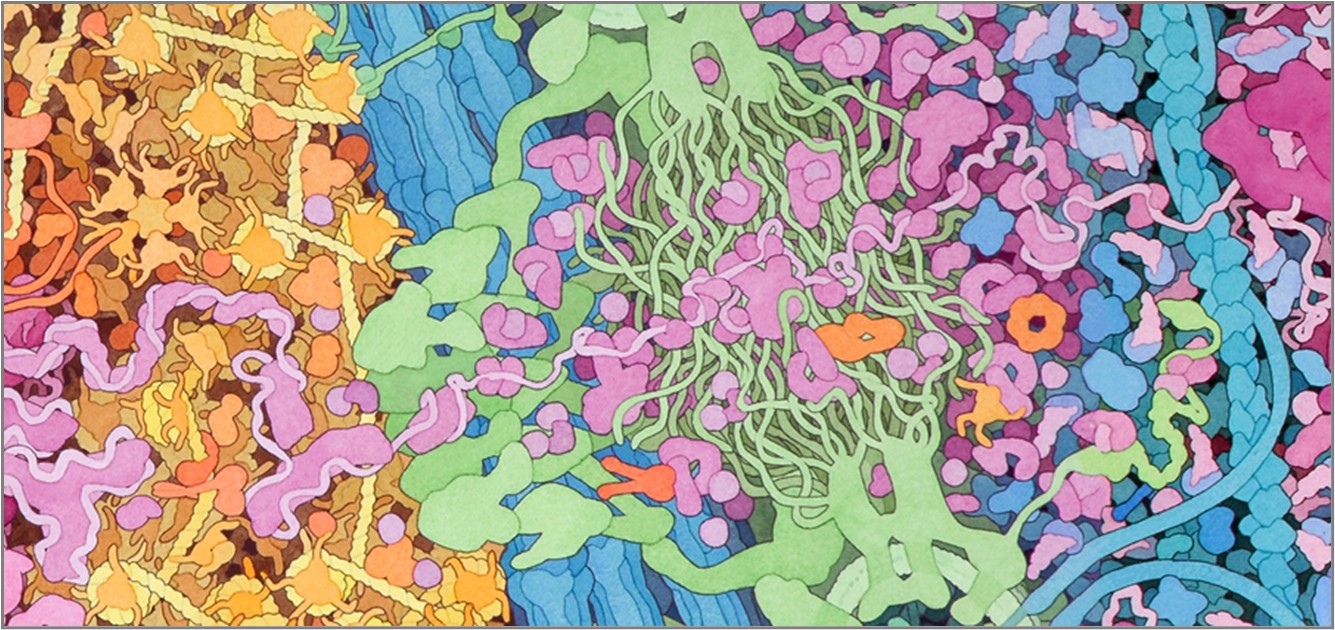
Both of the RNA subunits contain both ribosomal RNA (rRNA) and numerous ribosomal proteins. Below is the structure of a bacterial ribosome. Human ribosomes are even more complex, but share similar overall features! rRNA molecules are in bright colors, and ribosomal proteins are in soft colors. The ribosome is a huge complex, so be patient after you click the buttons - it will take a bit of time to load, but it is worth the wait!
All structures are shown in backbone (just one point for each amino acid or nucleotide, with a line connecting adjacent residues) and spacefill (showing all atoms at their appropriate size).
The next two buttons show the small ribosomal subunit. rRNA molecules are in bright yellow and ribosomal proteins are in pale yellow. This is an initiation complex, so a single tRNA molecule (orange) is in the P site. The mRNA is in green.
Small Ribosomal Subunit backbone PDB ID: 4v4fThe next buttons show the complete ribosome. The color scheme is the same as above, with large subunit proteins in light blue and large subunit rRNA molecules in a darker blue. Note that there are now two tRNA molecules in the structure, so this is just as the first and second amino acid of the growing peptide chain have been joined. These amino acids are not shown in the structure.
Ribosome backbone model PDB ID: 4v4jThe large and small subunits join to form a functional ribosome only when they attach to an mRNA. Each ribosome has three binding sites for tRNA. The P site (peptidyl-tRNA binding site) holds the tRNA carrying the growing polypeptide chain). The A site (aminoacyl-tRNA binding site) holds the tRNA carrying the next amino acid to be added to the chain. Discharged tRNAs leave the ribosome from the E site (exit site).
12. Does the 5' end or the 3' end of the mRNA strand attach to the small ribosomal subunit?
Slide your mRNA into the small ribosomal subunit. Refer to the photo below to ensure the mRNA is in the proper orientation in your ribosome. Keep sliding your mRNA into the small ribosomal subunit until the initiation codon of the mRNA is below the P site of the ribosome. Now attach the appropriate tRNA-amino acid complex to the mRNA in the P site. Note that translation of your mRNA will not begin at the very 5' end of the mRNA!
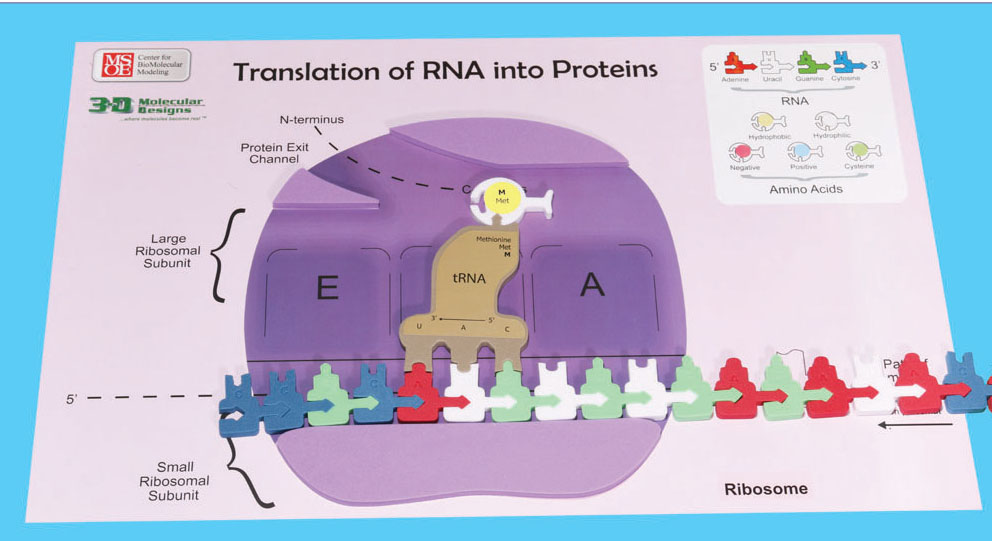
13. Which tRNA anticodon and accompanying amino acid will attach first in this P site?
In the next part of this activity you will model the elongation and termination processes of translation.
The anticodon of another tRNA base pairs with the mRNA in the A site. Complete this process using your model.
14. Which tRNA-amino acid complex will attach into the A site at this time?
An rRNA found in the large ribosomal subunit catalyzes the formation of a peptide bond between the amino group of the amino acid in the A site and the carboxyl end of the growing protein in the P site. The polypeptide is removed from the tRNA in the P site and attaches it to the amino acid still attached to the tRNA in the A site. Simulate the peptide bond formation with your model.
The ribosome translocates (shifts) the tRNA in the A site to the P site.
15. Circle and label the peptide bond in the photo below.
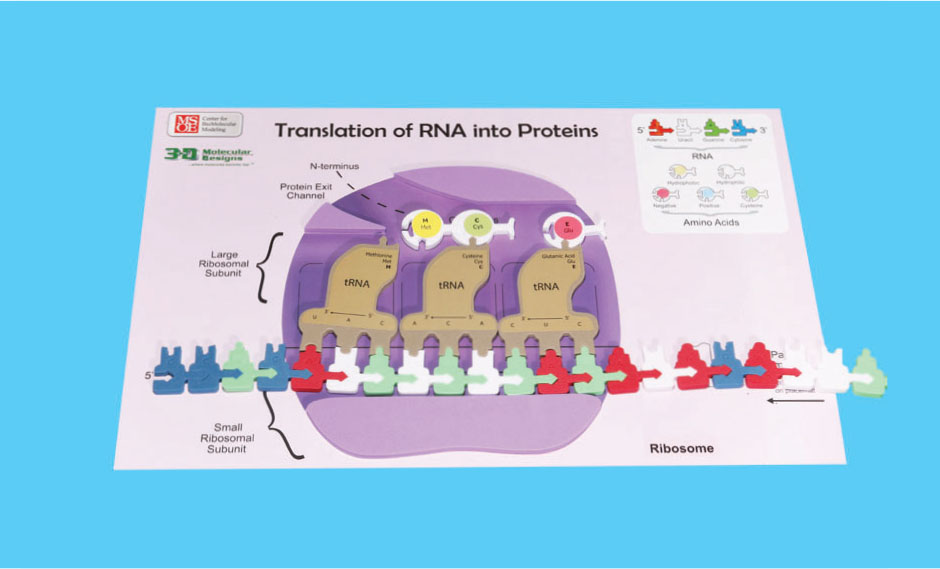
The tRNA in the P site is simultaneously moved to the E site where it is released.
Separate your tRNA in the E site from the mRNA and return the tRNA to the cytoplasm.
16. Which mRNA codon is now located in the A site?
With the A site now available for another tRNA-amino acid complex these steps can continue. Remember that the growing polypeptide transfers from the P site to the A site. Demonstrate this process using all of your tRNA-amino acid complexes in order.
The mRNA is translated in one direction from its 5' The mRNA is translated in one direction from its 5' → 3' end.
This developing polypeptide will exit the ribosome through the opening (exit channel) in the large ribosomal subunit.
A stop codon is also present to indicate the end of the protein. release factor binds to the A site, resulting of release of the protein and dissociation of the large and small ribosomal subunits and mRNA. Demonstrate termination of translation on your model.
17. Using the mRNA Codon/Amino Acid Chart list all stop codons.
18. What was the stop codon in the mRNA sequence that you translated?
19. What is the order of amino acids in your polypeptide? (Use the three letter and one letter abbreviations for each amino acid.)
20. When you reach the end of the mRNA strand describe what has happened to the polypeptide.
21. What will happen next to the polypeptide?
22. As you have followed this process of translation what steps are now left to be completed?
23. What will happen to the mRNA, tRNA, and the ribosome at the end of this process?
24. How long did this process of translation take for you and your lab group?
25. Do you think the cell could operate at this rate?
The mRNA, tRNA, and ribosomes can be reused over and over. The same protein can be made again if needed, or a new piece of mRNA can be translated. Ribosomes add new amino acids to the polypeptide at a rate of 20 amino acids per second (at 37° C).
26. At this rate, how long would it take to make a protein such as actin which is 375 amino acids long?
For a quick review of translation, view this stop motion video: