



This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
This activity explores protein primary and secondary structure. Buttons throughout the activity will generate protein structures in the Jmol window at the right. You may rotate molecules using 'click and drag' in the Jmol window.
This icon indicates you are to provide a response. You may enter your response in the activity, then download a file with your answers at the end of the tutorial. Alternately, you may download and print a paper copy of the worksheet.
Proteins consist of linear chains of amino acids that fold into specific shapes in 3D space. Twenty different amino acids are used in building proteins; amino acids share a common 'backbone' structure, but each has a different sidechain (sometimes called 'R' group).
The primary structure of a protein is the amino acids in order – like beads on a string. It is often designated with the single letter code of each of the amino acids. An example would be A-W-S-K which stands for alanine-tryptophan-serine-lysine. This information comes from the DNA, is transcribed by the m-RNA, and translated into a protein by the t-RNA and ribosomes.
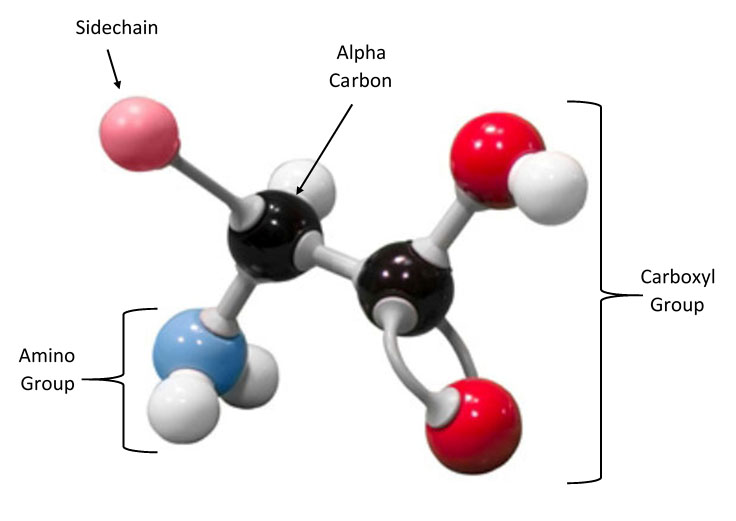
The backbone consists of an alpha carbon atom (the carbons are named using Greek letters, and alpha is the first letter of the Greek alphabet). The common backbone atoms in each amino acid include the alpha carbon attached to 1) an amino (NH3) group, 2) a carboxylic acid (COOH) group and 3) a hydrogen atom. The sidechain is also attached to the alpha carbon, but this is the part of the amino acid that varies.
Amino Acid StructureEach amino acid has the same backbone (NH2-CHR-COOH). The identity of the amino acids is determined by the chemical composition of the R group. We can group the sidechains according to chemical properties, such as hydrophobic (non-polar) versus hydrophilic (polar).
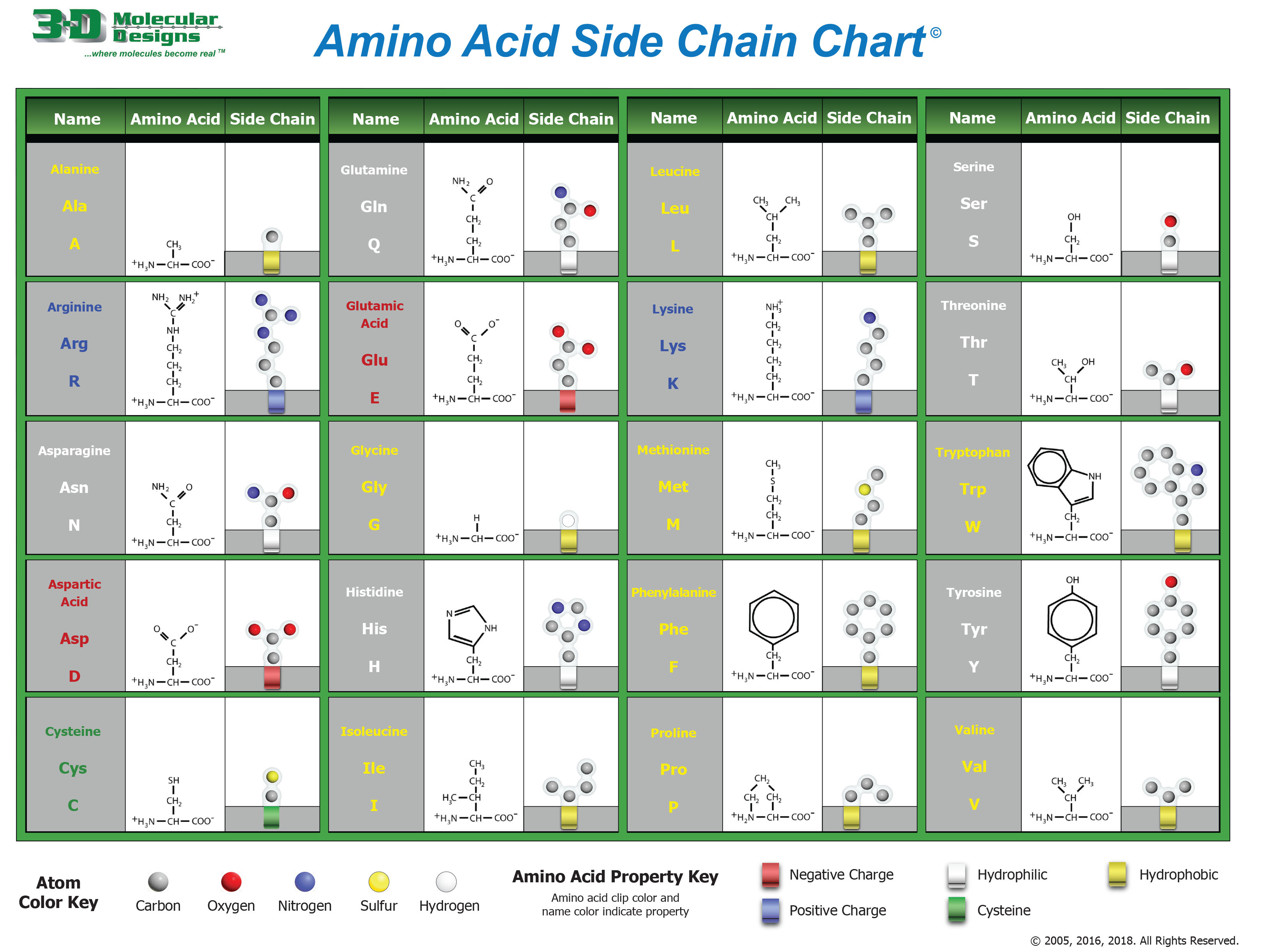
The chemical properties of the amino acid sidechains dictate how the protein folds in 3D space – since the sequence of amino acids is different for each protein, each protein folds in a unique way, and the protein shape and chemical properties determine the protein's function.
The secondary structure of proteins is due to the amide H (H of the nitrogen) of one amino acid making a hydrogen bond to the carbonyl O (C=O) of another amino acid. These are on the backbone of the protein. ONLY the protein backbone is involved in the formation of the secondary structure. The two common secondary structures are the alpha helix and the beta pleated sheet. In proteins these can be joined by loops of less regular protein structure. The alpha helix and the beta pleated sheet are pictured below.
Alpha helix carbon backbone with hydrogen bonds PDB ID: 1htmInterspersed among these secondary structural elements are loops, which are more flexible regions of the protein. Proteins can be composed of primarily α-helices (for example, β-globin) or β-sheets (for example, GFP), or a combination of the two structural motifs.
The following buttons illustrate the overall secondary structure of three proteins. The proteins are displayed in an alpha carbon backbone format. An alpha carbon backbone connects the alpha carbon atoms of consecutive amino acids, essentially creating a dot-to-dot 3D structure of the overall shape of the protein.
In each protein, helices are salmon, sheets are yellow and loops are white. The N terminus (amino group of first amino acid in chain) is colored blue, and the C terminus (carboxyl group of last amino acid in chain) is colored red. You can click and drag on the image to rotate each model.
Beta sheet carbon backbone with hydrogen bonds PDB ID: 1embThe primary sequence of a protein, composed of amino acids, determines the organization of the sequence into the secondary structure. There are two periodic secondary structure motifs, α-helix and β-sheet. Proteins can be composed of primarily α-helices (for example, β-globin) or β-sheets (for example, GFP), or a combination of the two structural motifs.
The α helix/β sheet construction kit will allow you to explore the characteristic properties of these two secondary structures. If you do not have access to the kits, the images and videos below will help you work through the activity.
The alpha carbon backbone of an amino acid has the structure: NH3-C-COOH. The bond angle of the peptide bond formed between adjacent amino acids differs in α helices and β sheets. There are two different types of backbone pieces in the kit to represent these bond angle differences. Backbone pieces that form an α helix have a green dot on the α carbon, and backbone pieces that form a β sheet have a yellow dot on the α carbon.


Atoms in the kit are colored using standard cpk coloring:

1. Draw the chemical structure of the backbone of an amino acid.
2. Identify the α-carbon with an arrow. What bonds to the α-carbon?
3. What part of the amino acid structure is missing in this representation?
Join the α helix backbone pieces by connecting the nitrogen (blue) of one amino acid to the carbonyl carbon (gray with red oxygen attached) of another amino acid to form a linear sequence. Recall that amino acids are joined by a condensation reaction that removes a water molecule (hydrogen atom from amide group and hydroxyl atom from carboxylic acid) to create the peptide bond. These models only depict the peptide bond and not individual amino acids, so you will not see the water molecule that is created.
 Alpha helix backbone atoms PDB ID: 1htm
Alpha helix backbone atoms PDB ID: 1htm4. Describe the stability/flexibility of the alpha helix backbone.
Now add the hydrogen bonds between the hydrogen on the nitrogen (hydrogen isn't shown, so connect to magnet on nitrogen atom) and the carbonyl oxygen (again, the C+O bond isn't shown, so add to the magnet on the oxygen). Note the white bead on the hydrogen bond; this bead represents the hydrogen atom, which is part of the amide (NH3) group. Hydrogen atoms are not typically shown in accurate physical models because 1) their actual location can't be determined in X-ray crystallography and 2) it is easier to see the underlying structure if hydrogen atoms aren't displayed.
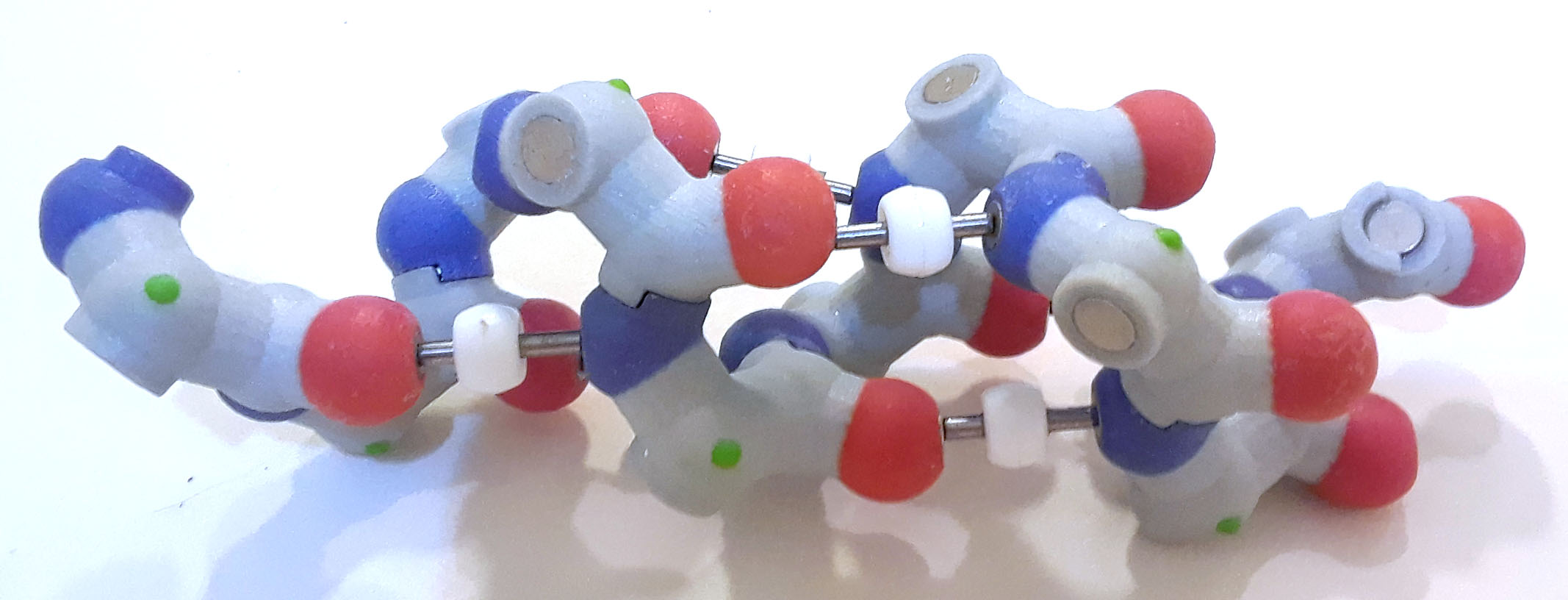 Alpha helix backbone atoms with hydrogen bonds PDB ID: 1htm
Alpha helix backbone atoms with hydrogen bonds PDB ID: 1htm5. How do the hydrogen bonds affect the structure of the alpha helix?
Now add the side chains to each of the alpha carbons.
 Alpha helix cpk backbone atoms with hbonds, sidechains in light blue PDB ID: 1htm
Alpha helix cpk backbone atoms with hbonds, sidechains in light blue PDB ID: 1htmThe following video shows rigid models of an alpha helix backbone and alpha helix with sidechains. The models are easier to rotate than the model with magnets.
6. Where are the side chains (R groups) – inside the alpha helix or outside the alpha helix? Why?
An alpha helix is a compact right-handed helix, with 3.6 amino acids per turn of the helix. The amino acid sidechains are bonded to the alpha carbon of each amino acid and radiate outward from the helix. The alpha helix is stabilized by hydrogen bonds – weak bonds between the amino nitrogen of one amino acid (x), and the carbonyl oxygen of another amino acid (x+4) located four residues further along the chain.
Using β sheet backbone pieces (yellow dot), build two short beta strands. Join the amide nitrogen of one amino acid to the carbonyl carbon of another amino acid to form a dipeptide, then continue to add backbone pieces to create two chains of five amino acids. Remember that in reality a water molecule is created during this reaction but is not shown in these models.

7. Do the two beta strands connect to each other?
We will be creating an antiparallel β sheet. This means that adjacent β strands point in the opposite direction. To do this, rotate one of the β strands, so that the nitrogen (N terminus) of one chain lies across from the carbonyl oxygen (C terminus) of the other strand. The N-C-C backbone of the two strands runs in opposite directions.
 Antiparallel strands backbone atoms PDB ID=1emb
Antiparallel strands backbone atoms PDB ID=1embNext, create a hydrogen bond between the backbone nitrogen on one strand with the carbonyl oxygen from the other strand. (Remember that the hydrogen shown in the bond is attached to the nitrogen, and the -OH from the carboxyl group was removed to create the peptide bond.) Add hydrogen bonds along the entire length, effectively 'stitching together' the two strands.
 Antiparallel beta strands backbone atoms with hydrogen bonds PDB ID: 1emb
Antiparallel beta strands backbone atoms with hydrogen bonds PDB ID: 1emb8. How do the hydrogen bonds affect the structure of the beta sheet?
You constructed an antiparallel β sheet, in which the N-terminal ends are at the opposite ends of the sheet. If two adjacent strands of a β sheet both have the N-terminal ends on the same end of the sheet, the strands are parallel. The N-C-C backbone of the two strands runs in the same direction.
 Parallel beta strands; N termini flash in green PDB ID: 1emb
Parallel beta strands; N termini flash in green PDB ID: 1embExplore the Jmol images below and compare the positioning of the hbonds in parallel and antiparallel beta strands.
Antiparallel beta strands backbone atoms with hydrogen bonds PDB ID: 1emb9. The hydrogen bonds in a (circle one) parallel / antiparallel sheet form a zig-zag pattern and the hydrogen bonds in a (circle one) parallel / antiparallel sheet are parallel to each other, like the rungs of a ladder.
10. Which type of sheet (parallel or antiparallel) do you think is more stable and why? The terms
The terms parallel and antiparallel refer to two adjacent strands in a β sheet, and a sheet with three or more strands can consist of a combination of parallel and antiparallel strands. Below is a section of a β sheet from green fluorescent protein (GFP, the protein that makes jellyfish glow) with strands color-coded. See if you can determine which strands are parallel and which are antiparallel.
Rainbow sheet backbone atoms with hydrogen bonds PDB ID: 1emb11. Give an example (using colors) of two adjacent strands that are antiparallel in GFP.
12. Give an example (using colors) of two adjacent strands that are parallel in GFP.
Add the sidechains to the amino acids in your beta sheet.
 Parallel beta strands backbone atoms cpk and sidechains lightblue PDB ID: 1emb
Parallel beta strands backbone atoms cpk and sidechains lightblue PDB ID: 1embThe following video shows rigid models of a beta sheet backbone and beta sheet with sidechains. The models are easier to rotate than the model with magnets.
13. Where are the side chains (R groups) on the beta sheet? Why?
A beta sheet is an extended, zig-zag structure in which individual strands are positioned parallel or anti-parallel to each other to form flat sheets in proteins. Since the amino acid sidechains are bonded to the alpha carbons of each amino acid, they are alternately orientated above and below the plane of the sheet. The beta sheet is stabilized by hydrogen bonds between the amino nitrogen's hydrogen atom of one amino acid and the carbonyl oxygen of another amino acid in an adjacent beta strand.