



This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
This tutorial explores how enzymes function, ranging from the enzyme-substrate interactions, enzyme specificity, cofactors and coenzymes, regulating enzymes and enzyme inhibitors.
All the enzymes explored in this tutorial have had their structures determined using X-ray crystallography or NMR. These structures are deposited in the Protein Data Bank (PDB) for use by researchers, educators and students. The PDB accession numbers are provided for each protein in this tutorial, in case you want to explore the proteins further.
Throughout the tutorial, clicking on the buttons will launch a Jmol image of the protein on the screen at the right.
Once a structure appears in the Jmol window, wait until the image stops moving before pushing a button to execute another script. You may spin the structure(left mouse button click and drag), or, if you wish, you may use Jmol commands to explore the structure further. A Jmol Quick Reference Sheet and Jmol Training Guide are available if you want to play with the images.
As you work through the tutorial, the question mark icon will signal that you need to answer a question. With some exceptions, these answers can be inserted directly into the Jmol Exploration, then emailed to your instructor by clicking the button at the end of the exploration. Alternately, you may download a copy of the worksheet.
Although the focus of this tutorial is on how enzymes function, all of the examples are important enzymes frequently discussed in biochemistry courses. You may explore these enzymes in greater detail throughout your career, so enzyme names and functions are included.
Enzymes are biological catalysts that make chemical reactions happen faster by offering an alternative pathway with lower activation energy. The enzyme is not changed in the chemical reaction but facilitates the reaction.
Below is what occurs. The enzyme (E) and substrate(S) noncovalently bind and change shape to form an enzyme-substrate complex (E-S). This is the binding step. The enzyme substrate complex then goes through a change to become the enzyme and product (P). This is the catalytic step.
E + S = ES → E + P
Why do the enzyme and substrate form a non-covalent bond rather than a covalent bond?
Enzymes are divided into groups — one method is by the type of reactions that they catalyze.
These groups are:
Oxidoreductases catalyze oxidation reduction reactions.
Transferases catalyze the transfer of a functional group from one molecule to another.
Hydrolases catalyze the addition of water and the breaking of a bond. These are often digestion reaction.
Lyases catalyze the addition of a group to a double bond or the removal of a group to make a double bond.
Isomerases catalyze the rearrangement of a functional group in a molecule or change isomers from one type to another.
Ligases catalyze the condensation (joining) of two molecules or the addition of carbon dioxide. Ligases do two different things to four different bonds (C-C, C-O, C-S, and C-N).
Enzymes and substrates fit together to form an enzyme-substrate complex. There are two models of this binding process, one is the lock and key and the other is the induced fit model. Neither is totally correct for all enzymes; they are only models to help us understand the behavior of the enzyme and substrate. (Remember that a model is a simplification of reality and does not fit all conditions and reactions).
One model is the lock and key model. In this model the enzyme and substrate fit together like a lock and key so that only the correct substrate will fit into the enzyme 'pocket' called the active site.
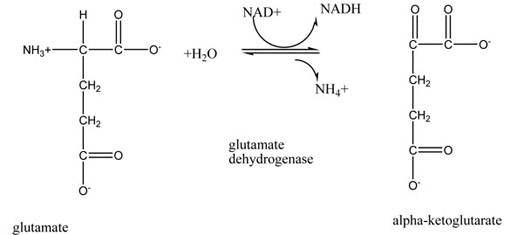
Click on the button below to see all the amino acids involved in the active site of the enzyme glutamate dehydrogenase. Glutamate Dehydrogenase (1BGV.pdb) is an enzyme that removes the nitrogen group (as ammonia) from the amino acid glutamate and makes alpha-ketoglutarate. Alpha-ketoglutarate is the carbon skeleton of glutamate and an intermediate in the Citric Acid Cycle. Glutamate dehydrogenase only can change glutamate, which fits perfectly into the active site.
Active Site PDB ID: 1bgvWhat color are the atoms that you see in the active site? Remember that they are in CPK colors - therefore - what atoms are there?
Structurally, what is the active site made of - what types of molecules are there?
The next button shows the substrate for this enzyme. The enzyme backbone is displayed in pale yellow, and the backbone of residues that interact with the substrate is colored pink.
Substrate PDB ID: 1bgvWhat atoms are present in the substrate?
The button below displays the substrate and sidechains of amino acids that interact with it in the active site of the enzyme. Amino acid sidechains are in 'soft cpk' colors, which are lighter hues of cpk colors. Soft cpk is used to distinguish enzyme atoms from substrate atoms. The substrate flashes in cyan, then returns to cpk coloring.
Active Site with Substrate PDB ID 1bgvThe reaction catalyzed by glutamate dehydrogenase is shown below:
In this model the substrate causes the enzyme to conform around it. HOWEVER – the substrate must be about the correct size for this to occur.
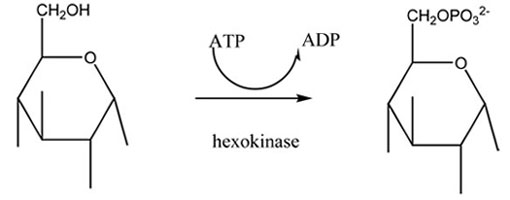
Hexokinase is the first enzyme in the glycolytic pathway, converting glucose to glucose-6-phosphate by adding a phosphate group. In the absence of substrate, the enzyme is in an 'open' conformation. When the substrate binds, the enzyme folds around the substrate in a 'closed' conformation.
The reaction is as follows:
Hexokinase is shown here in the open state, with no substrate bound.
Open State Hexokinase PDB ID: 3o80What atoms are in the active site?
Structurally, what is the active site made of - what types of molecules are there?
The next button displays the 'closed form' of hexokinase with the glucose molecule in the active site. At the end of the animation, only the substrate is displayed.
Closed State Hexokinase PDB ID: 3o8mWhat atoms are in the substrate?
Click the next button to watch the conformational change of hexokinase. This animation was made using the Yale Morph Server and the 3O80.pdb and 3O8M.pdb files. Rotate the protein during the animation to view the conformational change from all sides!
Conformational Change of HexokinaseWhat is the difference between the open and closed forms of hexokinase?
What atoms on the amino acids are coming together to close the glucose into the hexokinase?
What two possible interactions could be occurring here to have the hexokinase close?
The place that the enzyme and substrate interact is called the active site. This is often a depression or a pocket in the three-dimensional structure of the enzyme that at least part of the substrate fits into.
Do you think that the size and shape of the active site affect how the enzyme works? Why?
The active site consists of both binding and catalytic sites.
The binding site is where the substrate and the enzyme bind via non-covalent interactions.
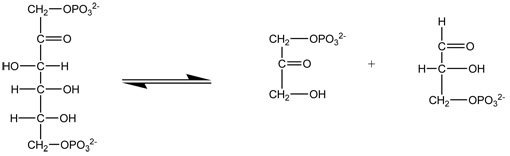
Fructose 1,6-bisphosphate aldolase catalyzes the reversible reaction of converting fructose 1,6-bisphosphate into dihydroxyacetopne phosphate (DHAP) and D-glyceraldehyde 3-phosphate (G3P). This is a step in the glycolytic pathway. In this representation of fructose 1,6-bisphosphate aldolase, the binding site flashes in yellow and soft cpk, then the substrate flashes in cyan and cpk. The substrate is displayed at a smaller scale to help distinguish it from the enzyme.
Binding Site PDB ID: 4aldLocate the amino acid residues that are involved in binding the substrate.
What part of those amino acids is interacting with the substrate?
If the enzyme and substrate formed covalent bonds, would they be difficult to separate?
What part of the enzyme binds to the substrate in general? Remember that enzymes are proteins.
What do you think is the purpose of the binding site?
What types of noncovalent interaction could be occurring?
The active site of Fructose 1,6-Bisphosphate Aldolase also has catalytic groups. Their shape is complimentary to the shape of the substrate. The catalytic groups are amino acid R groups.
What do you think is the function of the catalytic groups?
The catalytic groups of this enzyme are Asp33, Lys146, Arg148, Glu187, Lys229, and Arg303. They are shown flashing in lime green and cpk in the image below. Binding residues are in soft cpk. Locate catalytic groups in the image.
Catalytic Group PDB ID: 1aldBoth the binding site and the catalytic groups are R groups of amino acids. Do you think that the type of R group (hydrophobic, polar neutral, positive, or negative) has an impact on the jobs of the binding site and the catalytic groups? Why?
Write a complete sentence describing what occurs at the binding site and the catalytic groups of an enzyme's active site.
Enzymes are specific to particular reactions. There are four types of specificity – absolute, group, linkage, and stereochemical. Not all enzymes work on all substrates.
Enzymes with absolute specificity can act upon one and only one substrate. The enzyme glutamate dehydrogase (1BGV.pdb) will catalyze only the removal of the nitrogen group from glutamate- NOT any other amino acid. It has absolute specificity.
Active Site PDB ID: 1bgvWhat atoms/groups are in the active site of this enzyme?
What atoms are in the substrate?
What possible interactions could be occurring between the substrate and active site?
Why do you think that - speculate - this enzyme has absolute specificity based on the structure that you see?
What would be an advantage and a disadvantage of absolute specificity?
Group specificity means that an enzyme will catalyze a reaction on a function group of a variety of molecules (for instance, cleaving an alcohol group or working only on aldohexose sugars).
The enzyme sulfur oxygenase reductase (SOD) catalyzes the reduction of inorganic sulfur compounds and elemental sulfur to sulfate and is a major part of the global sulfur cycle. This enzyme has a group specificity of only sulfur compounds. This is a very large protein with 24 subunits that form a sphere when assembled. Images below only explore a portion of protein, but you will see the curvature of the 'leaflet' and should envision leaflets assembling into a sphere.
Protein Overview - SOD PDB ID: 2cb2The chemical reaction catalyzed by sulfur oxygenase reductase is as follows:
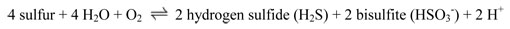
Here the active site for the sulfur substrate is composed of a cysteine persulfide (flashes lime green) and an iron atom (flashes light blue) held by a 2-His-1-carboxylate facial triad (the CPK amino acids). In this case, the 2-His-1-carboxylate facial triad is composed of 2 histidines and 1 glutamate. Note that each identical subunit contains an identical active site.
Active Site SOD PDB ID: 2cb2What atoms of the enzyme/amino acid residues are probably holding the cysteine and persulfide?
Let's explore the active site in more detail. There is not a substrate pictured in the active site; the researchers are not yet able to crystallize the substance.
Active Site in Detail PDB ID: 2cb2The iron is necessary for this reaction to occur. Looking at the name of the enzyme and remembering what you learned in general chemistry - what type of reactions are occurring in this enzyme?
What would be an advantage and a disadvantage of group specificity?
Enzymes with linkage specificity are able to break or form a single type of bond.
Here we explore the active site and the substrate of fructose 1,6-bisphosphate aldolase (4ALD.pdb), an enzyme that is part of the glycolytic pathway. The substrate is fructose 1,6-bisphosphate, and a specific bond is broken to produce dihydroxyacetone phosphate and glyceraldehyde-3-phosphate.
What atoms/groups are in the active site of this enzyme?
What atoms are in the substrate?
What possible interactions could be occurring between the substrate and the active site?
Below is the reaction catalyzed by fructose 1,6-bisphosphate aldolase.
Click the next button to see which bond is cleaved to produce the product.
Reaction Site PDB ID: 4aldGive another example of linkage specificity that we learned about when studying carbohydrates. (There are 2).
What would be an advantage and a disadvantage of linkage specificity?
Enzymes with stereochemical specificity are able to catalyze a reaction with only one enantiomer of a pair. For instance, some enzymes can break down D-sugars but not L-sugars. Here we explore D-lactate dehydrogenase with the substrate D-lactic acid. The substrate flashes in cyan.
Active Site PDB ID: 3kb6Do you think that the other enantiomer, L-lactic acid, would fit in this enzyme's active site? Why or why not?
Do you think there are a lot of these types of enzymes? Why?
What would be an advantage and a disadvantage of stereochemical specificity?
Write a complete sentence for each type of specificity.
Some enzymes need cofactors or coenzymes to function properly. Many coenzymes are made of vitamins, so they are organic molecules. Many cofactors are minerals, so they are metal ions or atoms. Vitamins and minerals are necessary for good functioning of the body.
Cofactors are often found in the active site of enzymes and change the shape of the active site, thus allowing the substrate to bind to the enzyme. Without the cofactors, the enzyme is non-functional. Here we explore enolase. The substrate of the enzyme appears in light CPK colors, and the two metal ions flash in yellow.
Enolase with Cofactor PDB ID: 2oneWhat metal atoms are the cofactors in this enzyme? (Hint: you must scroll your mouse over the atom to find its designation)
Do you think that this enzyme could function correctly without the metal ions? Why or why not?
Coenzymes work in a different method. Coenzymes usually move electrons or other functional groups from the substrate to somewhere else. Examples of coenzymes are the electron carriers NAD+/NADH and FAD/FADH2. In this image of glyceraldehyde-3-phosphate dehydrogenase (3KSZ.pdb) the turquoise-flashing molecule is NAD+, a coenzyme that takes hydrogen ions from the substrate and changes to NADH. The NADH then delivers hydrogen ions to the electron transport system (ETS) to make ATP. Note that the NAD+ is in the active site.
Coenzyme PDB ID: 3kszLocate the 3-phosphoglycerate and the NAD+. What atoms of the NAD+ are near what atoms of the 3-phosphoglycerate? Make an educated guess about what is occurring here.
What are two major differences between cofactors and coenzymes?
Write a complete sentence describing cofactors and coenzymes.
All organisms regulate their enzymes' actions. If organisms don't regulate their enzymes, they waste the precious energy (ATP) and would probably die.
Human regulation of enzymes is different than for bacteria. The types of regulations are below.
Allosteric control of enzymes means that another molecule (called an effector molecule) will bind to the enzyme at a site that is different than the active site and either turn the enzyme on (positive allosterism) or turn the enzyme off (negative allosterism).
Aspartate transcarbamoylase catalyzes the first step in the synthesis of pyrimidines. The positive effector molecule that turns this enzyme on is ATP, which flashes in lime green. This is positive allosterism. Note that ATP DOES NOT BIND in the active site. The active site is shown briefly in orange spacefill at a completely different location in the protein.
Positive Allosterism PDB ID: 4at1What do you think the positive allosteric effector molecule - in this case ATP - is doing to the active site of the enzyme to turn it on?
This same enzyme also is turned off by CTP, which flashes in turquoise (5AT1.pdb). The CTP binds to the same area that the ATP bound, but turns the enzyme off. This is negative allosterism. Again note that it DOES NOT bind in the active site (shown in orange).
Negative Allosterism PDB ID: 5at1What do you think the negative allosteric effector molecule - in this case CTP - is doing to the active site of the enzyme to turn it off?
Write a sentence about positive allosterism.
Write a sentence about negative allosterism.
In feedback reguation, the final product of series of enzymatic reactions will turn off the first step. This occurs because the final product is accumulating, and further production would consume substrates and energy. When levels of the final product decline, less product is available to bind to the enzyme, and synthesis of the product resumes.
Here the A-F are substrates and the E1-E5 are the enzymes that catalyze each reaction.
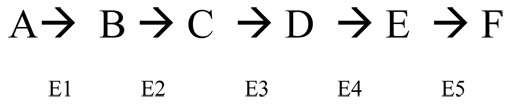
Which molecule (substrate) will turn off the first reaction's E1 if this enzyme works by feedback inhibition?
The enzyme aspartate transcarbamoylase is the first enzyme in a series of reactions that makes CTP (5AT1.pdb). This enzyme is turned off by CTP (which fades in and flashes in turquoise) in a feedback mechanism because the CTP is the final product and if there is enough, the enzyme will be turned off. (Note that CTP functions as a feedback regulator through negative allosteric control. Categories of enzyme regulation are not always mutually exclusive!)
Feedback Inhibition PDB ID: 5at1Another way that organisms regulate enzymes is by using a proenzyme (or zymogen). The enzyme initially has extra amino acids attached that cause it to be inactive. This often happens with digestive enzymes – so they don't destroy the producer cells. When they get to the place where they will do their work, the extra amino acids are removed and the enzyme becomes active.
Below is an example of what happens. The letters represent amino acids.
Inactive form of enzyme (proenzyme or zymogen) - portion that will become active enzyme is in red:
A-V-M-F-T-N-Y-C-K-R-H-E-S-G-I-W-L-M-N-Q-Y-C-S-S-S-R-H-D-E
Active form of enzyme:
A-V-M-F-T-N-Y-C-K-R-H-E-S-G-I-W-L-M-N
Pepsin is one of a series of digestive enzyme that break down proteins by cutting them into smaller peptides. It has a large active site pocket that can fit protein chains. Unless you have just eaten a meal, you don't want pepsin active in your digestive tract, as it will break down YOUR proteins. It exists in an inactive proenzyme, pepsinogen, that is activated by cleaving off the N terminal end of the protein. In the images below, pepsin has a light yellow backbone. The N terminus of the protein is blue and the C terminus is red. The sequence that is found in pepsinogen and cleaved to make the active protein is in green.
Proenzyme Pepsinogen PDB ID: 3psgWhat part of the above example is missing from the active form but present in the inactive form?
Would those missing amino acids have an effect on the final enzyme structure? Why or why not?
What parts of the molecule (what colors) are missing from the pepsin that are present in the pepsinogen?
Is the shape of the pepsin and pepsinogen the same or different? Why?
The last way that organisms regulate enzymes is by protein modification. A chemical group is added or removed from the enzyme and that either activates or deactivates the enzyme. Most commonly the group added is a phosphoryl group. This often occurs to enzymes of irreversible reactions that are in opposing pathways.
Below is a diagram of what occurs between the opposable enzymes of glycogen synthase (MAKES glycogen) and glycogen phosphorylase (DEGRADES glycogen). Note that in one case the enzyme is active with the phosphate and in the other case is inactive with the phosphate.
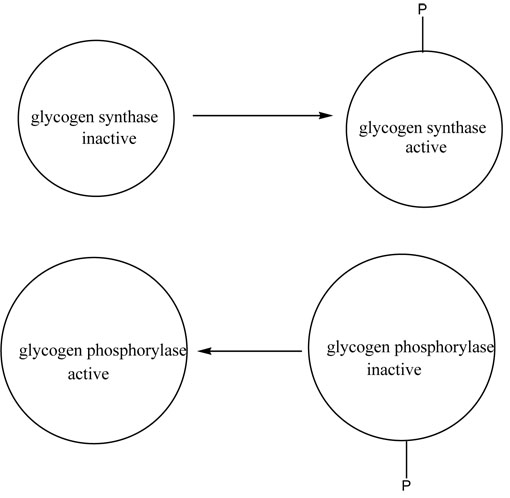
This enzyme shown below is glycogen phosphorylase. The active protein (8GPB.pdb) contains phosphate in the form of AMP which flashes in lime green, whereas the inactive protein does not have phosphate (9GPB.pdb). In both cases, the substrate flashes briefly and then is colored in light purple. Rotate the molecule to see how tucked away it is in the active site!
Active Protein with Phosphate PDB ID: 8gpbWhat would be an advantage of having the same group activate some enzymes and deactivate others?
Write a complete sentence describing each type of enzyme control that humans use to control their enzymes.
Drugs and poisons are chemicals from the environment that often affect enzymes. This is done deliberately in the case of prescription drugs and things like aspirin, etc. These chemicals are called inhibitors if they decrease the catalytic action of the enzyme. They are called activators if they increase the catalytic action of the enzyme. NOTE – THESE ARE NOT THE SAME AS THE AFFECTOR MOLECULES THAT TURN ON AND OFF ALLOSTERIC ENZYMES. These outside chemicals are either reversible or irreversible inhibitors. The reversible inhibitors are either competitive or noncompetitive inhibitors.
Irreversible enzyme inhibitors bind very tightly to the enzymes, often by covalent bonds. This can affect the enzyme by having a change in shape of the enzyme, binding in the active site and blocking any substrate, or interfering with the catalytic groups of the active site so that the substrate can't be changed to the product.
Some examples are arsenic which binds to the thiol group (SH) of cysteine and causes a shape change in enzymes. Snake venom, nerve gas, and penicillin are also examples of this type of inhibition. These types of chemicals often inhibit many enzymes.
This enzyme is acetylcholinesterase (1CFJ.pdb), irreversibly bound to Sarin, a nerve agent. Acetylcholinesterase recycles the neurotransmitter acetylcholine so it can be used again to transmit nerve signals. Sarin flashes in turquoise. Rotate the molecule to see exactly how Sarin is bound.
Acetylcholinesterase with Irreversible Inhibitor Sarin PDB ID: 1cfjWhy do irreversible inhibitors have to form a covalent bond to the enzyme to stop the enzyme from functioning?
Write a sentence defining irreversible inhibition of enzymes.
Reversible competitive inhibition is done by molecules called structural analogs. These molecules are close in structure and charge distribution to the substrate. When drug companies are looking for new drugs to alter biochemistry (for example treating depression) this is often what they want. The inhibitors occupy the active site but can't be changed to the product. They compete with the substrate for the active site. So, obviously the concentration of substrate and inhibitor is important. If there is more substrate, the enzyme works. If there is more inhibitor, the enzyme doesn't work. This enzyme is dihydrofolate reductase (3DFR.pdb), bound to methotrexate, a drug used to treat a variety of diseases, including cancer. The methotrexate flashes in turquoise.
Reversible Competitive Inhibitor PDB ID: 3dfrIf you were designing another drug like methotrexate to do the same type of thing, what type of structures would you look at to design this new drug?
Does a competitive inhibitor have to be the exact same size as the substrate - or does just part of it have to be the same? Why or why not?
Do the inhibitor and the substrate compete for the active site?
What would be the effect if excess inhibitor was added?
What would be the effect if excess substrate was added?
Reversible noncompetitive inhibitors usually bind to one of the groups in the active site or to a metal ion cofactor. The binding is weak, usually noncovalent, and the enzyme activity comes back when the enzyme and the inhibitor dissociate from the Enzyme-Inhibitor Complex (E-I complex). Some inhibitors don't bind in the active site, but elsewhere and cause a shape change to occur in the enzyme. This also inactivates a wide range of enzymes.
Shown here is hexokinase, complexed to glucose, which flashes in turquoise, and the inhibitor, glucose-6-phosphate, which flashes in lime green (1BG3.pdb). Rotate the enzyme to see the positions of the substrate and inhibitor!
Rotate the model to see the relative positions of substrate and inhibitor.
Do the inhibitor and the substrate compete for the active site?
What would be the effect if excess inhibitor was added?
What would be the effect if excess substrate was added?
Write a complete sentence describing each type of inhibition.