

This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
The structure of DNA was first determined in 1953. This exercise will explore the structure of DNA, beginning with nucleotide structure, through hydrogen bonding of base pairs and the structure of the sugar-phosphate backbone to the DNA double helix.
Models in this activity are from the DNA Discovery Kit. If you have the DNA Discovery Kit, you can work through the activity with the models. If you don't have access to the DNA Discovery Kit, the images will help you complete the activity. Buttons will execute Jmol animations related to DNA structure to the right. These images may be rotated by positioning your mouse over the image and using a 'click and drag' motion.
This icon indicates you are to provide a response. You may enter your response in the activity, then download a file with your answers at the end of the tutorial. Alternately, you may download and print a paper copy of the worksheet. Question numbers are included to allow you to easily transition from the tutorial to the worksheet.
If you have access to models, compare the parts in each of the four plastic bags. Otherwise, compare the parts in the images below.




1. What parts of all nucleotides are the same?
2. What parts of all nucleotides are different?
Each nucleotide consists of three parts:
These models are based on X-ray crystallographic structures and are built to scale. X-ray crystallography does not permit the identification of hydrogen atoms, so these are not shown on the model, except in the case of hydrogen bonds between base pairs. These are shown in white and are held together using magnets. The remaining colors used in the model are standard CPK colors, as follows:
Click on the buttons below to view the structures. You may use your mouse button to rotate the image (click and drag) or zoom (shift, click and drag). Hovering over an atom will identify it.
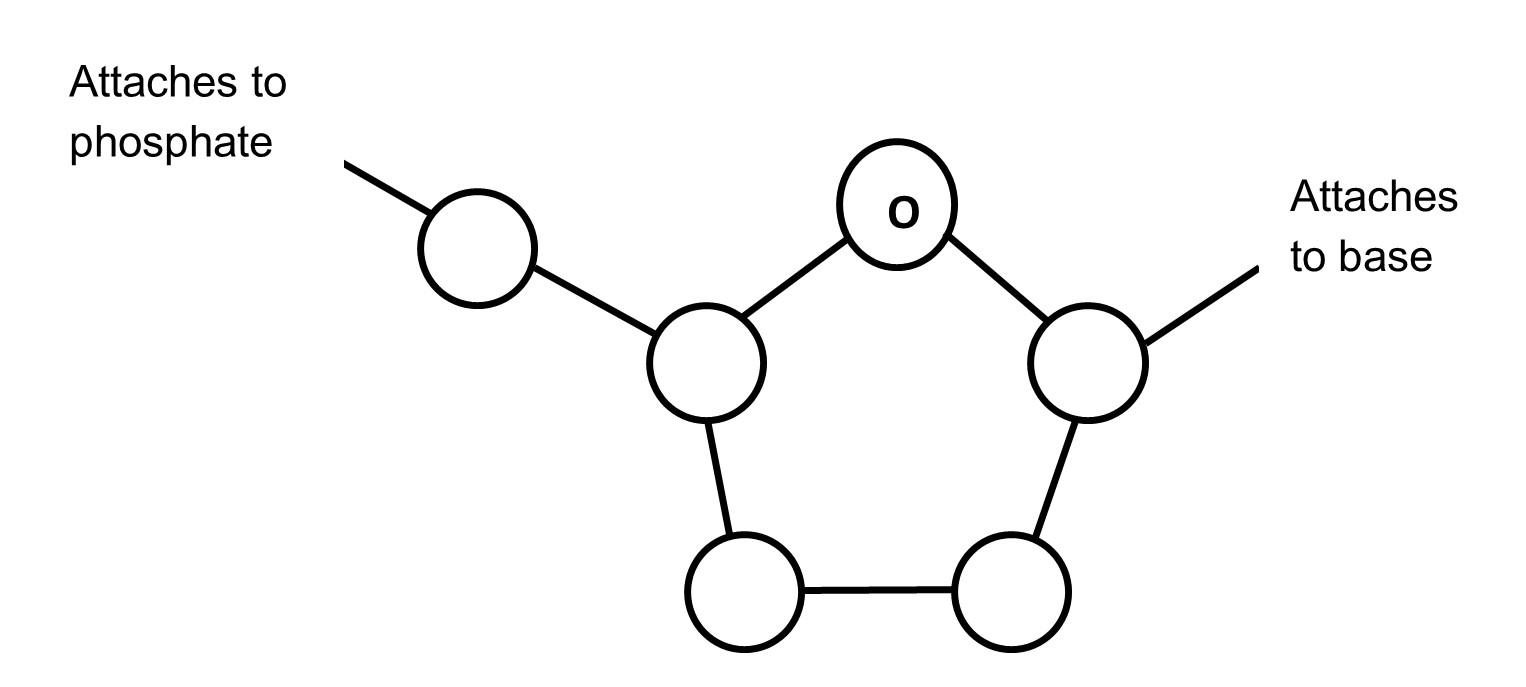
phosphate group3. Below is the structure of the deoxyribose. Number the carbons (1' to 5') and place an asterisk (*) on the carbon that contains an -OH group in ribose. [Numbering begins with the carbon that attaches to the base.]
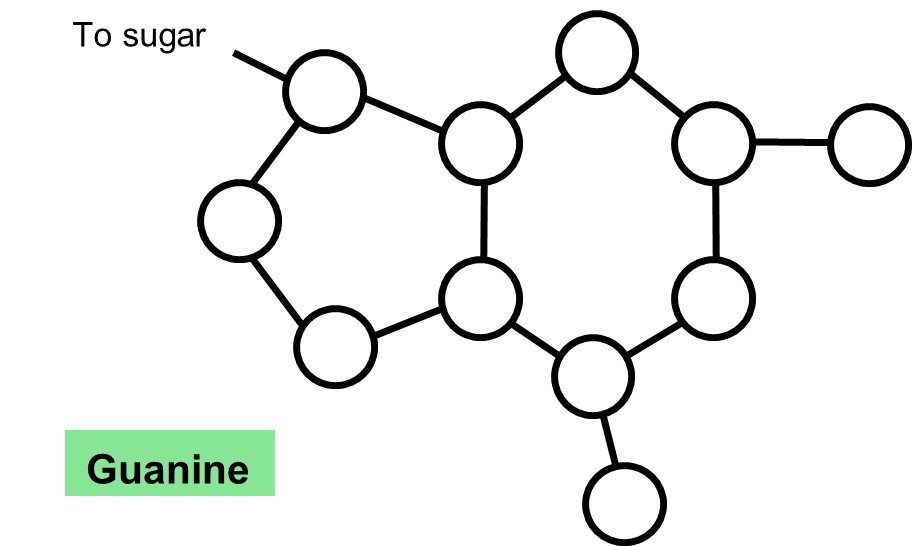
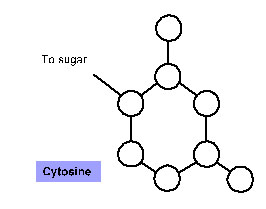
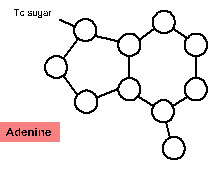
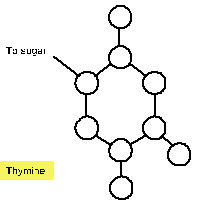
4. For each of the nitrogenous bases below, label the atoms as carbon (C), oxygen (O) or nitrogen (N). Notice that Hydrogens (H) are not shown on the diagrams below. Draw dotted lines to indicate where the hydrogen bonds form with the complementary base. NOTE: you cannot draw hydrogen bonds to connect the complementary bases.
If you have access to models, assemble each nucleotide using only the pegs and holes. Remember, each nucleotide consists of a sugar group to which are attached a phosphate group and a nitrogenous base.
Next, determine which bases are most likely to hydrogen bond together. Remember that the width of the DNA helix is the same for each base pair. This can only be achieved with a purine, which contains two ring structures, pairing with a pyrimidine, which contains one ring structure. Note that in order for the bases to hydrogen bond, one of the bases in each pair in the diagrams above must be flipped upside down.
5. Based on your drawings, record your prediction as to which bases pair with which: __________ pairs with _________ and __________ pairs with _________ .
Test your hypothesis by aligning the base pairs in the model. When done correctly, the magnets (representing hydrogen bonds) will hold each base pair together.
base pair 1; hydrogen bonds joining base pair in green6. Indicate which of the bases (A, C, G and T) goes with each of the following terms. Some descriptions will require more than one answer.
______ purines
______ pairs with C
______ has three hydrogen bonds
______ pyrimidines
______ pairs with G
______ has two hydrogen bonds
______ pairs with A
______ pairs with T
______ attaches to the 1' carbon of deoxyribose
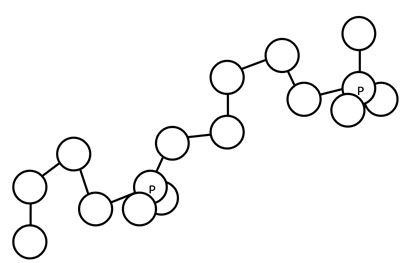
Nucleotides join together in a chain, with the hydroxyl group on the 3' carbon of one sugar attaching to the phosphate group on the 5' carbon of the sugar on the adjacent nucleotide. If you have access to the models, join the phosphate group of one nucleotide with the sugar group of another nucleotide (they snap into place when they are in the correct orientation), then rotate the structure so the bases are in the back and parallel to the table.
sugar-phosphate backbone (atoms not displayed in drawing in soft cpk colors)7. Label the atoms that you see in the sugar-phosphate backbone. Circle the 5' carbon of each deoxyribose. Place an asterisk next to the 3' carbon of each deoxyribose. Finally, draw an arrow to indicate the direction, from the 5' end of the molecule toward the 3' end of the molecule.
If you are working with models, assemble the base pairs into a double helix. Assemble the helix on the stand. Your final model will look similar to the DNA below, although the arrangement of nucleotides may differ.
DNA8. Using either the model or the Jmol image of DNA, sketch the order of the bases on the helix and indicate the 5' and 3' ends of the strands. It may be useful to draw the bases on one strand upside down, to remind you that two strands run in opposite directions.
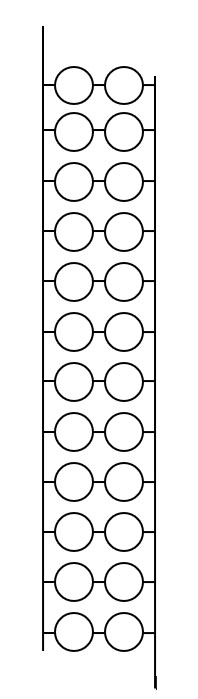
9. Explain what is meant by the term 'antiparallel.'