



This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
The SARS-CoV-2 virus is part of the coronavirus family of viruses (Astuti & Ysrafil, 2020). The SARS virus is an airborne virus that is spread by saliva droplets (Astuti & Ysrafil, 2020). Specifically, SARS-CoV-2 is responsible for the COVID-19 pandemic that is continuing to impact many people worldwide, having caused over 2 million deaths. The virus can cause mild flu-like symptoms to severe illnesses (Viner et al., 2020).
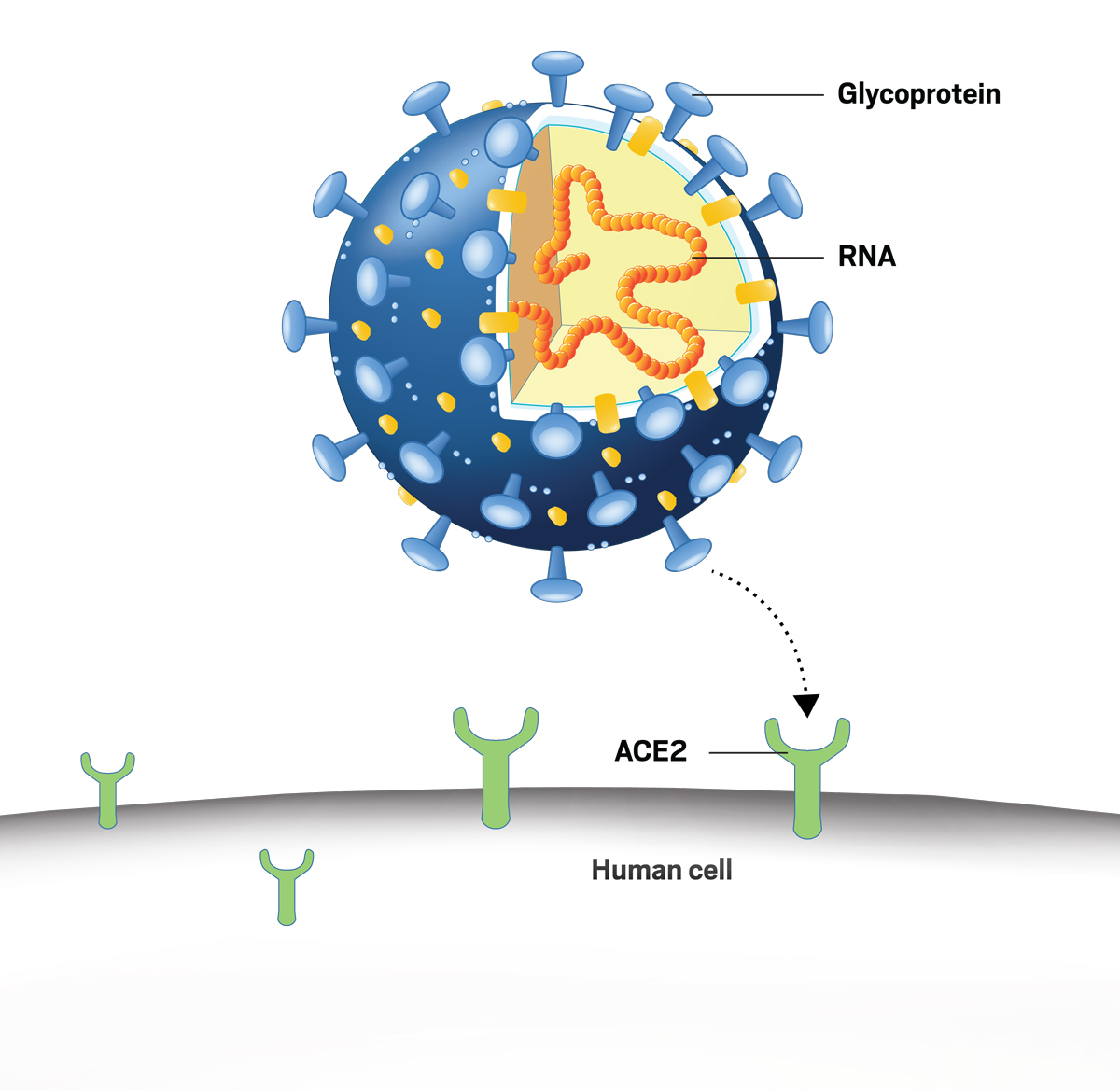
When the virus enters the body, it uses its spike proteins (glycoprotein) to attach to healthy human cells via the ACE2 receptor (Ni et al., 2020). After binding with the ACE2 receptor protein, it fuses with the human cell and is then able to reproduce (Ni et al., 2020).
To neutralize the virus, monoclonal antibodies bind to the epitopes of the SARS-CoV-2 spike proteins. By doing this, the antibodies block the spike proteins from binding to the ACE2 receptors on human cells (Wang et al., 2020).
The monoclonal antibodies 80R and 362 are known to bind to the epitopes on the spike protein receptor-binding domain (RBD) to neutralize the virus (Walls et al., 2020). The 80R antibody binds to SARS-CoV-1 and the MAb362 antibody can bind to both SARS-CoV-1 and SARS-CoV-2. Likely, MAb362 can bind to both viruses since it has smaller amino acids in its binding domain. This can lead to more room in the molecular region where the antibody binds to spike protein. This characteristic will be investigated in the rest of the tutorial.
Below is an image that depicts the connectivity and effectiveness of 80R, MAb362, and NSU1 (new hypothetical antibody, more information in next section) antibodies to neutralize both the SARS-CoV-1 and SARS-CoV-2 viruses (Manikkuttiyil et al., 2021).

Click on the two Jmol buttons (gray bars) below to view the protein database files for the antibodies 80R and MAb362.
80R Antibody bound to SARS-CoV-1 spike protein (2GHW) (Hwang et al., 2006)Protein visualization software was used (Pymol and Jmol) to hypothesize a potentially more universal antibody called NSU1. NSU1 was expected to bind more effectively to future mutations in the SARS spike protein.
At the binding interface, between these antibodies and the SARS spike protein, MAb362 mutations trend smaller and less polar: Arg149Ser, Asn151Ser, Asp170Gly, and Trp213Ser. Due to the mutation trends presented by Mirsky et al., it was hypothesized that more space in this area could lead to increased resistance to future SARS-CoV spike protein structure variations (2015).
NSU1 was modeled based on MAb362 with the following four additional mutations: Asp103Gly, Trp104Leu, Gly170Ser, and Arg211Val (Manikkuttiyil et al., 2021).
In the image, MAb362 has more space in the binding domain (circled in red) because of the smaller amino acids compared to 80R. The extra space allows for better connectivity and effectiveness in neutralizing the virus. The hypothesized antibody NSU1 continues to follow this trend.

Click on the Jmol buttons (gray bar) below to view the hypothesized antibody NSU1.
NSU1: Hypothesized Antibody (modified from MAb362) (Manikkuttiyil et al., 2021)The table below provides a summary of the various amino acid changes in each of the models. The colors in the table match the colors in the models.
Three differences between the spike protein residues of SARS CoV1 and CoV2 were found (Manikuttiyil et al., 2021). Nine amino acid residues were found to be different between 80R and MAb 362, 3 between the heavy chains, and 6 between the light chains. Four additional amino acids (highlighted in purple) were selected from MAb 362 to change into a hypothetical and more universal antibody shown in NSU1 (Manikkuttiyil et al., 2021).

We would like to thank the Center for BioMolecular Modeling (CBM), who granted a travel and education grant to E. Schmitt Lavin. A special thanks to Margaret Franzen, Ph.D. and Mark Hoelzer of the CBM for their guidance on this project. In addition, we want to thank CBM for their encouragement in making 3-D molecular modeling more accessible and cost-effective for students, scientists, professors, and physicians in the healthcare realm. We are appreciative of the CREST program for travel awards and making this project possible. We would also like to extend a special thank you to the lab of Dr. Celia Schiffer and Shurong Hou, Ph.D. who provided us with the MAb362 pdb file.
Astuti I, Ysrafil. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes Metab Syndr. 2020;14(4):407-412. doi:10.1016/j.dsx.2020.04.020
Ejemel, M., Li, Q., Hou, S. et al. A cross-reactive human IgA monoclonal antibody blocks SARS-CoV-2 spike-ACE2 interaction. Nat Commun 11, 4198 (2020). https://doi.org/10.1038/s41467-020-18058-8
Hwang WC, Lin Y, Santelli E, Sui J, Jaroszewski L, Stec B, Farzan M, Marasco WA, Liddington RC. Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J Biol Chem. 2006 Nov 10; 281(45):34610-6. https://doi: 10.1074/jbc.M603275200
Manikkuttiyil, C., Abbas, F., Abbas, L., Alzamora, C., Hunt, M., Julakanti, P., Likki, S., Manikkuttiyil, C., Schmitt Lavin, E., Sikora, A. Minor Changes with Large Implications: Modeling Amino Acid Mutations in SARS-CoV Monoclonal Antibodies (80R and 362) Towards the Design of More Universal Antibodies. Experimental Biology, 2021. Abstract R2150.
Mirsky A, Kazandjian L, Anisimova M. Antibody-Specific Model of Amino Acid Substitution for Immunological Inferences from Alignments of Antibody. Molecular Biology and Evolution, 2015 Dec 21; 32(3): 806–819. https://doi.org/10.1093/molbev/msu340.
Ni, W., Yang, X., Yang, D. et al. Role of angiotensin-converting enzyme (ACE2) in COVID-19. Crit Care 24, 422 (2020). https://doi.org/10.1186/s13054-020-03120-0
Viner RM, Ward JL, Hudson LD, et al. Systematic review of reviews of symptoms and signs of COVID-19 in children and adolescents [published online ahead of print, 2020 Dec 17]. Arch Dis Child. 2020;archdischild-2020-320972. doi:10.1136/archdischild-2020-320972
Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020 Apr 16; 181(2): 281-292.e6. doi: 10.1016/j.cell.2020.02.058.
Wang, C., Li, W., Drabek, D. et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun 11, 2251 (2020). https://doi.org/10.1038/s41467-020-16256-y