

This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
Remdesivir (GS-5734) is an inhibitor of the viral RNA-dependent RNA polymerase (RdRp) which has shown inhibitor capabilities against severe acute respiratory syndrome coronavirus (SARS-CoV-1) and the Middle East respiratory syndrome (MERS-CoV) in vitro. Because of its ability to inhibit SARS-CoV-2 in vitro, remdesivir was initially believed to have potential therapeutic uses against SARS-CoV-2 and received Emergency Use Authorization from the FDA. However, trials after the authorization found that remdesivir was ineffective in decreasing the mortality rate. This could be since the intact viral genome copies still may be assembled, escape the cell, and help the virus proliferate.
RNA-dependent-RNA-polymerases (RdRps) are enzymes that catalyze the synthesis of new RNA. The RdRp is essential for almost all RNA viruses because it is what these viruses use to replicate their genomes and, therefore, is highly conserved across RNA viruses, which makes it a convenient drug target. RdRp binds to a template strand of RNA and will take nucleosides from the environment to build a complementary strand.
The RdRp of SARS-CoV-2 is specifically made up of three different non-structural proteins (nsp): nsp7, nsp8, and nsp12. Nsp12 is the largest of these, and its active-site cleft binds to the RNA, mediates the RdRp activity, and replicates the RNA genome. Recent structural analyses show a second nsp8 bound near the active site which extends down along the RNA up to twenty-eight base pairs away. The two copies of nsp8 bind to the opposite side of that cleft near nsp7. These are thought to confer processivity to nsp12 and allow for the protein to effectively interact with RNA. Helical nsp8 poles extend along the RNA and form positively charged 'sliding poles' that the RNA is guided along. These poles are thought to be what help replicate the virus's long genome. This second nsp8 will not be shown in our model due to the PDB structure used not having it included.
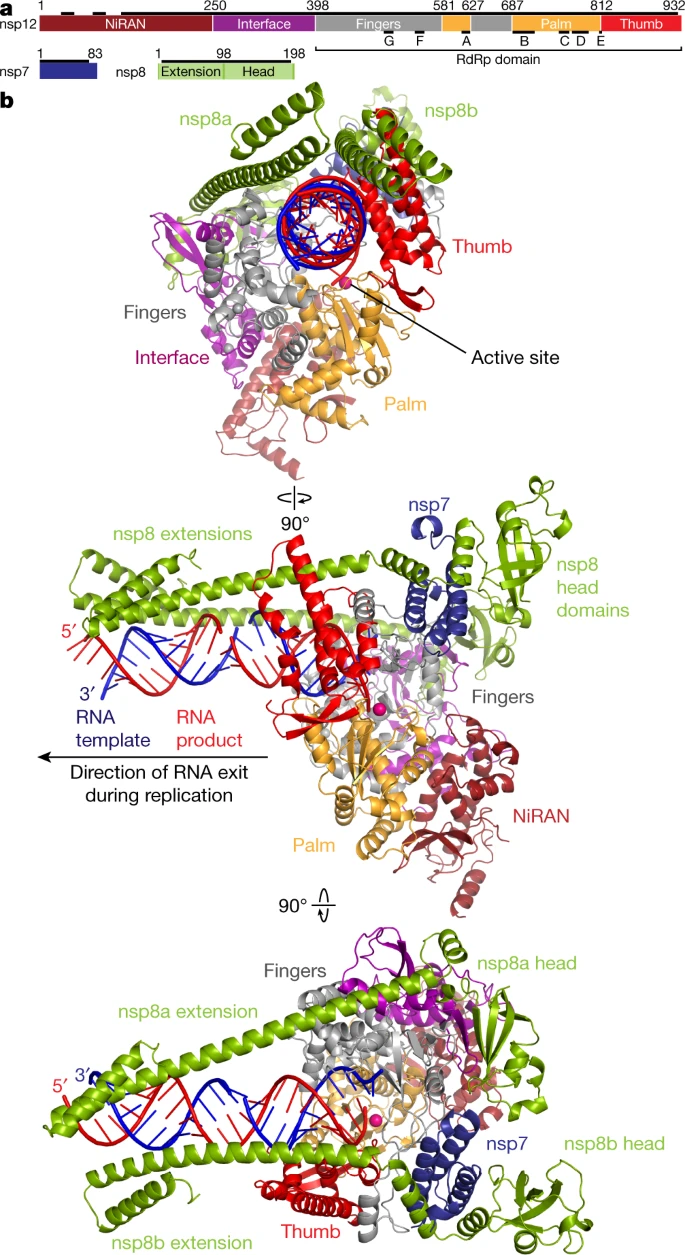
In our model, nsp12 is represented by a light green protein backbone, along with nsp8 being light cyan, and nsp7 being a light purple.
The RNA has a sandy brown backbone, along with the different RNA nucleosides being color-coded in a pastel version of their traditional color code. The model is in spacefill closer to the active site so that interactions between specific atoms and groups may be seen more easily.
At the active site, a CPK remdesivir molecule can be seen bound in place of an RNA nucleoside. This will be explained further in the following section.
Remdesivir is a nucleoside analogue. Nucleoside analogues resemble the nucleosides cytidine, thymidine, uridine, guanosine, or adenosine, all of which are necessary components of DNA and RNA. A nucleoside analogue can be used to 'trick' viruses and bacteria into using the false nucleosides instead of actual nucleosides, but due to the structural differences between the nucleosides and the analogues, their ability to properly replicate is inhibited. Nucleoside analogues also tend to be delivered to the body in a prodrug form, meaning that the drug isn't in its active state when injected into the body. The prodrug must be modified in the body to become its active drug state and then be structurally similar to the nucleoside analogue it resembles.
Remdesivir is recognized by viral organisms in its active state as adenosine due to their similar structures. This causes the virus to incorporate remdesivir into its genetic material and then relies on three central modifications to successfully stop the replication of viral RNA. These modifications cause instabilities that cause a termination of the RNA replication 3 base pairs late.
Comparison of adenosine and remdesivirRemdesivir differs from the structure of adenosine because of a cyano group (CN) attached to the 1' carbon of the ribose sugar. This changes the shape of the sugar, which then changes the shape of the RNA strand. It is theorized that this modification is the primary reason that RNA replication is terminated.
Remdesivir also has a carbon-carbon bond between the sugar and the base of the molecule rather than the carbon-nitrogen bond in adenosine. This is important because coronaviruses have an enzyme called exonuclease that recognizes unnatural nucleosides. However, with this modification, the remdesivir isn't recognized by the exonuclease. This modification can be seen in the diagram below.
