



This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
The SARS-CoV-2 virus, which causes the COVID-19 disease, has been the focus of concern around the world, setting records as a modern-day pandemic. The World Health Organization (WHO) has confirmed over 111 million cases in the world with over 2.5 million confirmed deaths as of February 23, 2021. SARS-CoV-2 continues to be a serious threat to global public health, leaving a distinct footprint in human history. Our research visualizes and contextualizes the mechanism of remdesivir, an antiviral nucleotide analog prodrug. Remdesivir targets the viral RNA-dependent RNA polymerase (RdRp) to inhibit replication of the virus. This process is facilitated by a subunit replication-and-transcription complex of three major nonstructural proteins (nsps): nsp7, nsp8, and nsp12. With our three-dimensional model (PDB ID 7BV2), we depict the mechanism of remdesivir inhibition of the copying of the viral RNA template through the central channel of RdRp by terminating chain elongation. When deciding the project, remdesivir showed promise as an inhibition drug for SARS-CoV-2, as it was effective for MERS-CoV. The National Institutes of Health (NIH) and the WHO have conflicting results about the effectiveness of remdesivir for treating COVID-19. Further comparison between the RdRp structures of SARS-CoV-2 and MERS-CoV may elucidate why remdesivir may be ineffective and could lead to better drug design. In our RNA model, the hydrogen bonds between the RNA primer strand and remdesivir are highlighted to show how remdesivir acts as a nucleotide to inhibit further viral replication. In the full RdRp complex, the hydrogen bonds between remdesivir and key residues of the nsp proteins are depicted. Our model illustrates the viral RdRp of SARS-CoV-2 and how remdesivir should impact viral RNA replication through inhibition.
At the beginning of 2020, the world was impacted by the SARS-CoV-2 virus and the COVID-19 disease, taking the lives of millions of people. For previous coronaviruses, such as SARS-CoV-1 (MERS-CoV), the antiviral prodrug remdesivir was effective in treating patients. Initially, remdesivir was promising as a therapeutic drug for SARS-CoV-2 as well. This led to our interest in the structure and function of remdesivir for SARS-CoV-2. The SARS-CoV-2 virus is composed of four structural proteins and sixteen non-structural proteins (nsps) that each have a function associated with viral function of the virus. The nsp7, nsp8, and nsp12 compose the complex containing the RNA-dependent RNA polymerase (RdRp), which is vital for transcription and replication of the virus. The nsp12-nsp7-nsp8 complex allows the template RNA strand to be replicated. Remdesivir targets RdRp by closely resembling the nucleotide, adenosine, and attempts to inhibit further elongation of the primer strand (chain P).
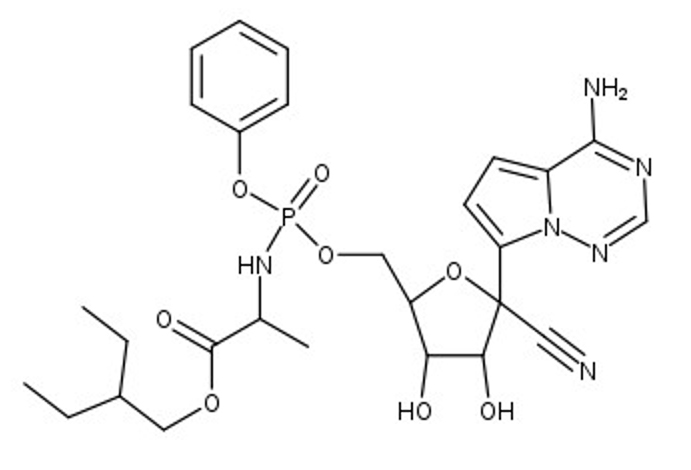
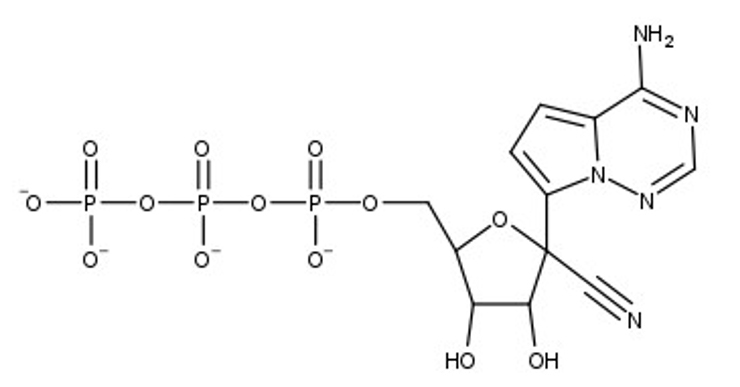
1. nsp12 is highlighted, notice the RNA primer and template are coming out of the nsp12.
2. nsp8 is highlighted as part of the RdRp complex.
3. nsp7 is highlighted as the final part of the active complex.
RdRp is rotated to show the entire complex. We, then, zoom into the active site and highlight remdesivir in green.
We zoom into the active site to see the hydrogen bonding (shown in yellow) in between the amino acid residues and remdesivir.
1. Notice remdesivr with thr 687 blinking in cyan and orange red.
2. lys 545 blinking in magenta and white.
1. Remdesivir colored in cpk, blinking in lawn green.
2. Attached to the light steel blue primer and hydrogen bonded (shown in yellow) to two oxygens in the template (lavender).
This class has been a different experience compared to what a typical course would look like. In the first semester, we had a few assignments that prepared us for the project by giving background knowledge to different topics, such as enzymes, amino acids, proteins, etc. These lectures were helpful to get an understanding about the material that we would need for our research. We believe this course had a very positive environment. Students didn't have to be stressed about exams but instead focus on learning by exploring and trial and error.
1. Biomolecular Modeling Course
- Students on the project were enrolled in general chemistry or organic chemistry
- Fall 2020 focused on background information (Proteins, Enzymes, Viruses, Jmol (coding), PDB)
- Electronic notebooks were kept with annotated documents
- Learned to read and understand peer-reviewed journal articles
- No tests were given
- Encouraged teamwork, making connections, and communication
- Poster abstracts were submitted to Experimental Biology (Apr 24-27 - Virtual) and PDB 50 (May 4-5 Virtual and Presenters are invited guests)
2. Educational attributes from 3-dimensional model and Jmol poster
- Hydrogen bonding
- 3-D structures of protein complexes
- RNA replication
- RNA dependent RNA polymerase in corona viruses
- RNA replication inhibition
Yin, W., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. doi.org/10.1126/science.abc1560