

This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
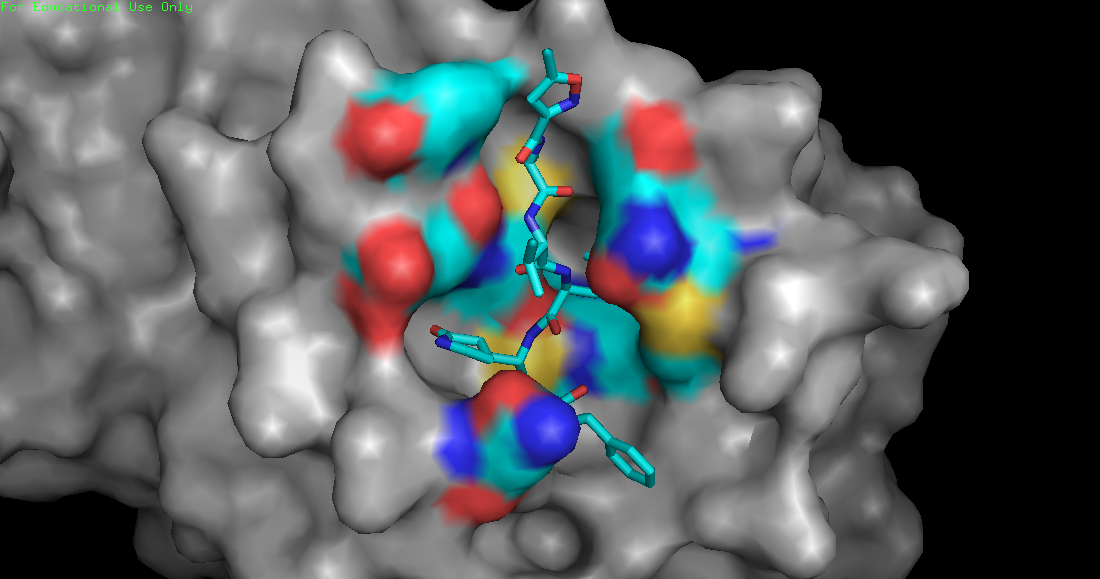
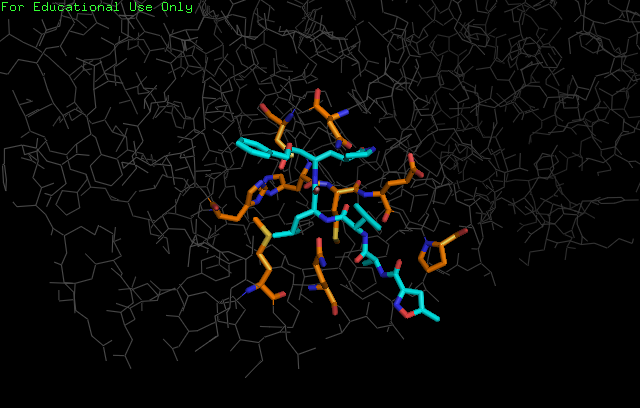
A new coronavirus causing an acute respiratory syndrome (SARS-CoV-2) emerged in late 2019 and has been responsible for the outbreak of coronavirus disease 2019 (COVID-19). Symptoms of COVID-19 range from mild to severe, including respiratory symptoms, systemic inflammatory responses, and even death. Currently, remdesivir is the only FDA-approved therapeutic agent to treat COVID-19, although it shows limited efficacy. We were interested in learning about the drug design process for a novel virus. We focused on drug research targeting the SARS-CoV-2 main protease, known as nsp5 or Mpro. The nsp5 protease is an enzyme critical for viral replication, with ebselen, cinanserin, and N3 being lead compounds identified as possible effective drugs (Jin, 2020). Starting with these chemical drug structures, we used an in silico drug design process to explore how to modify and refine these drugs. Our goal was to make the compounds more drug-like, according to Lipinski's and Veber's rules. Using the crystal structure file 6YB7 from the Protein Data Bank (PDB), we focused on the protease active site, defined by amino acids His41, Met49, Asn142, Cys145, His164, Met165, Glu166, Pro168, and Gln189. Cys145 is especially important, as it is the target for irreversible covalent inhibitors. Based on this information, we used molecular docking software to first re-dock these drugs into the nsp5 protease. We then made changes to the structures of the lead compounds, docked those new compounds, and used the energetic information to continue the refinement process. Once we completed our own drug design explorations, we created a three-dimensional printed model of the full nsp5 protease, with the key amino acids highlighted. We also created a three-dimensional model zoomed in on the active site, with the different drugs fitting into that active site model. Here, we present what we learned about the drug design process for a novel virus such as SARS-CoV-2. Our docking protocols and models are useful for teaching the fundamentals of drug design and about modern drug discovery and design processes.
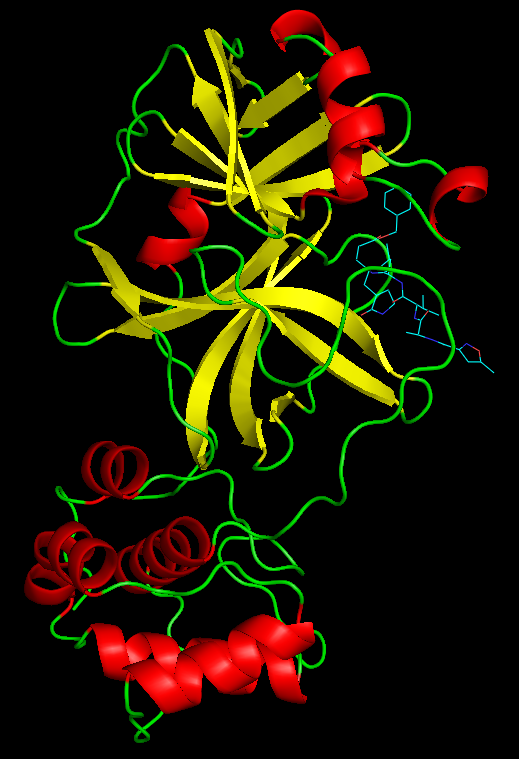
SARS-CoV-2 is part of the coronavirus family that has been responsible for the most recent global pandemic. Infection with the virus causes symptoms ranging from mild respiratory symptoms to fatality. The only drug approved to treat infection with SARS-CoV-2 is remdesivir. We were interested in exploring how additional new drugs could be developed to treat infected patients.
We chose to focus on the main viral protease, nsp5. Using the protein data bank file (PDB) 6yb7 of the SARs-CoV-2 nsp5 protease, we identified key amino acids that would help us when docking and manipulating a novel drug. Drug design was performed, exploring how drug design principles such as Lipinski's rule of 5 are relevant. The final PDB structure we worked with, 6lu7, was used for testing how well the drugs bound the nsp5 protease. Molecular docking was used to assess how well the newly designed drugs bound to the 6lu7 structure of the nsp5 protease.
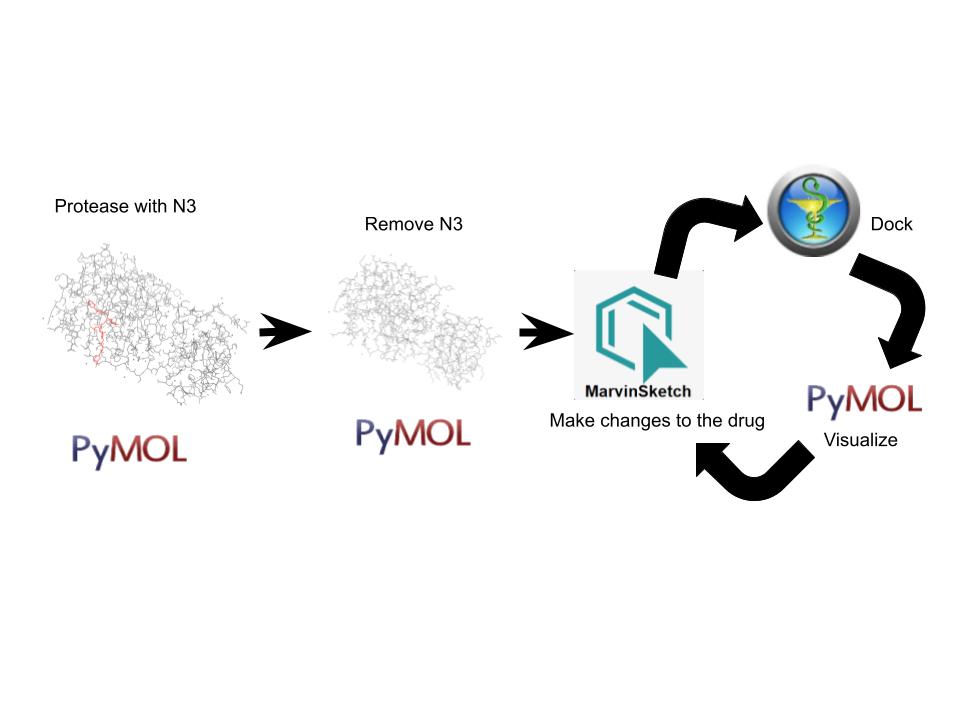
We used the drugs cinanserin, ebselen, and N3, to learn about the drug design process. We used an in silico docking method to dock them and gather information about how they bound within the main protease. We used the following software applications: Jmol, MarvinSketch, PyMOL, and PyRx.
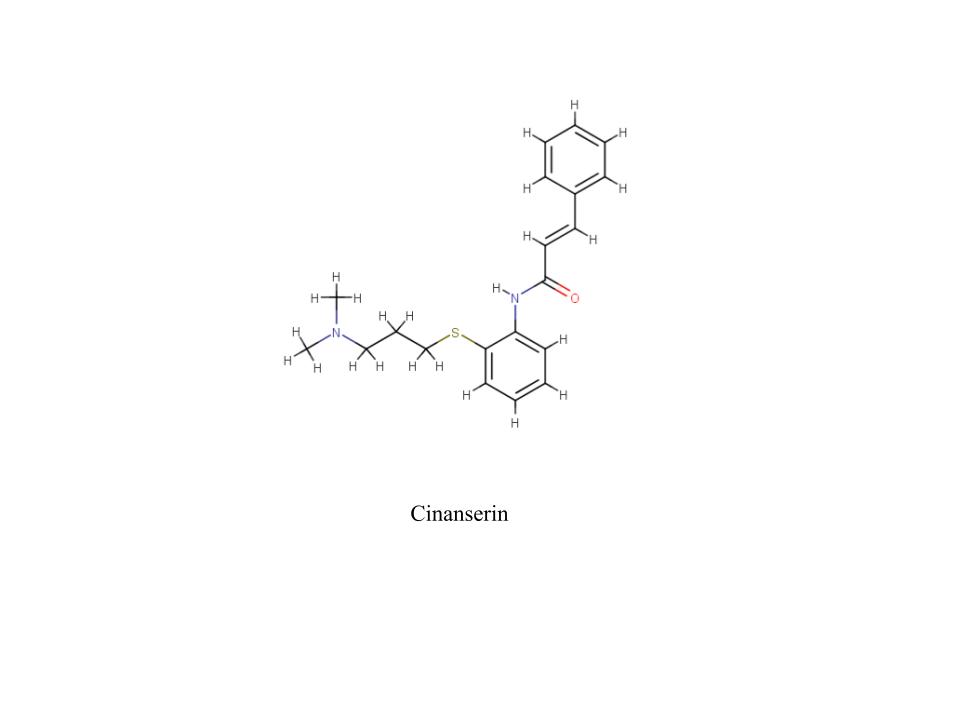
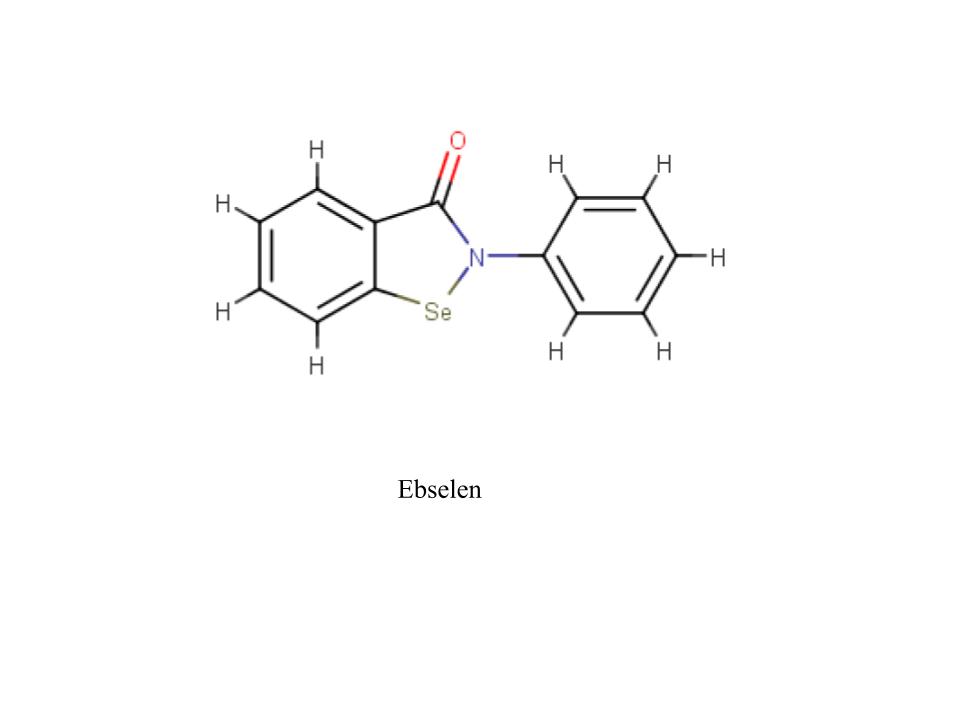
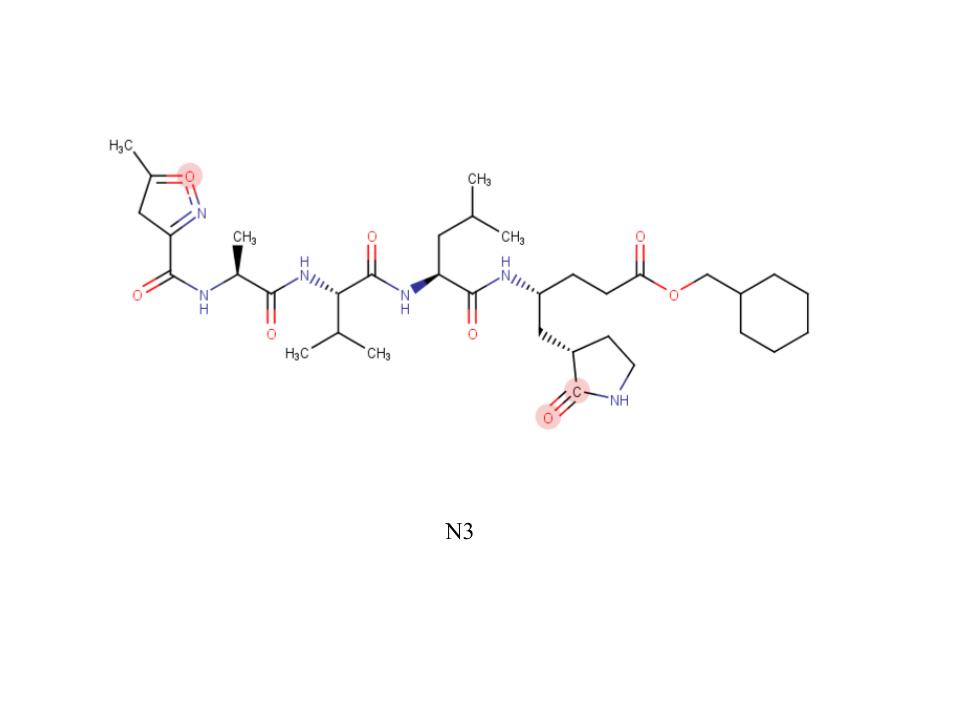

This course has not only provided useful information, but has also provided skills we will be able to use in future classes and even in our careers. Our confidence in applied learning, the scientific method, and academic independence has grown exponentially. In this course, we were given opportunities to explore the depths of the virus and the process of creating a drug to stop infection. Through this, we have gained experience in the areas of biology, organic chemistry, biochemistry, medicinal chemistry, and even molecular docking, coding, and scripting. In more depth, we spent time dealing with thermodynamics, functional groups, amino acids, line drawings, intermolecular forces, and more. As a group, we have improved our scientific dialogue and are confident we can carry these conversations with ease. The struggle of the rigorous material has shown to be worth it and we are proud of the work we have accomplished.

Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Jin, Z. et al. (2020) Nature 582: 289-293.
COVID-19 main protease with unliganded active site. Owen, C.D. et al. To be published.