This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
Succinate Dehydrogenase is found in the inner mitochondrial membrane, but a portion lays in the mitochondrial matrix. As shown in the Jmol diagram, the red represents the hydrophobic regions of the enzyme. As seen, there is an abundant of red at the top of the the enzyme. This is because these subunits lie in the inner mitochondrial membrane which is nonpolar. The 'bottom' or the other end of the the enzyme is covered in more blue, which is the hydrophobic regions, which lies in the mitochondrial matrix.
 Hydrophobic/Hydrophilic regions
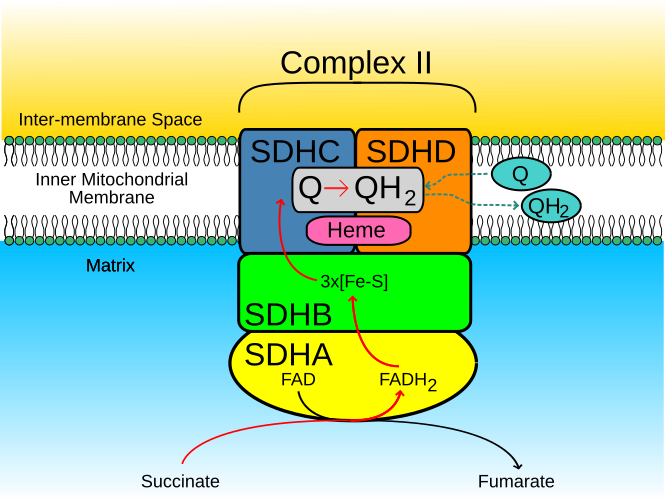
Hydrophobic/Hydrophilic regionsSuccinate Dehydrogenase functions in cell respiration, energy generation, oxygen level sensing, and tumor suppression. It participates in both the the electron transport chain and the citric acid cycle. In the electron transport chain it reduces ubiquinone to ubiqunol simultaneously as it oxidizes succinate to fumarate in the citric acid cycle.

Succinate Dehydrogenase contains four subunits.
The Four Different SubunitsSuccinate Dehydrogenase contains alpha-helices and anti-parallel beta- sheets. All of the beta-sheets are located in subunit A and B, while aplha-helices are located in all four subunits.
Alpha-Helix and Beta-sheetsLigands are non-proteins associated within an enzyme and amino acid side chain on a protein enzyme that interacts with enzyme and substrate. In subunit A there are three ligands, Flavin-Adenine Dinucleotide, (limegreen), Calcium ion, (red), and Oxaloacetate ion, (blue). FAD in this subunit is used as a cofactor for the substrate binding site in the electron transfer from succinate to fumarate in the citric acid cycle.
Ligands: Subunit AIron-Sulfur Proteins subunit, aka subunit B, consist of three Iron Sulfur clusters. One of which interacts with a Methionine through hydrogen bonding interactions. These clusters surround the substrate binding site which is connected by redox reactions suggesting physiological electron transfer.
Iron-Sulfur ClustersThe cytochrome b-556 subunit, aka, subunit C, consist of three ligands, Protoporphyrin, aka heme,L-Alpha-Phosphatidyl-Beta-Oleoyl-Gamma-Palmitoyl-Phosphatigylethanol amine, aka EPH, and Cardiolipin. EPH interacts with a Lysine through hydrogen bonding interactions.
Ligands: Subunit CThe Hydrophobic membrane anchor, aka subunit D, contains one ligand, Ubiquinone-2.
Ligand: Subunit DIn subunit A, the ligand FAD is used as a cofactor for the substrate binding site in the electron transfer from succinate to fumarate in the citric acid cycle.
Oxidation of Succinate to FumarateIn subunit B, C, and D, closes to the Iron-Sulfur Cluster (3Fe-4S)(red) in subunit B, a physiological quinone-buinding cite is near. The essential residues for ubiquinone-binding, which takes place in the electron transport chain when ubiquinone is reduced to ubiquinol is the heme b ligation which is located in a hydrophobic space created below the heme (blue). Subunits C and D (orange, green), form a membrane-bound cytochrome b with six transmembrane helices. In this site there is also a phospholipid molecule that has two acyl groups called a cardiolipin (yellow).
Ubiquinone-Binding Site