

This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
Citrate Synthase is the first enzyme found in the Krebs cycle. This enzyme is responsible for the condensation of acetyl-CoA and oxaloacetate to form citrate.

The overall structure for citrate synthase is two subunits where there are two active sites. Each subunit makes one citrate. The two subunits can be seen in the image below:

The top image shows the subunits with their product in red. The bottom image shows the subunits with their substrate in green.
For this enzyme, the product is going to be citrate. The substrate is oxaloacetate along with Acetyl-CoA and H20.
To see a closeup, general structure of one subunit, please click on the button below.
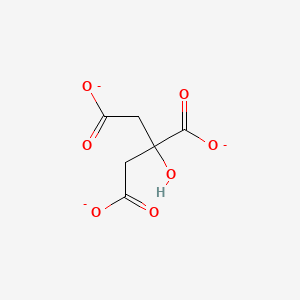
The structure of citrate synthase can be zoomed into citrate. Looking in this area, you are able to see the active site of the enzyme. Click on citrate to see this.
CitrateAs stated before, the substrate for Citrate Synthase is oxaloacetate (along with Acetyl-CoA and water). The reaction starts when oxaloacetate binds to the binding site in citrate synthase at amino acids His274, His320, and Asp275. This makes the enzyme go into its closed form. This means that Citrate Synthase is an induced fit enzyme. At this point, Arg329 forms a salt bridge with oxaloacetate. In its closed form, Acetyl-CoA now has access to the binding site. After a few reactions, there is an end product in which CoA is released and citrate is produced. Citrate synthase then changes back to its open form.
* As stated before,since there are two subunits, there are two active sites, which means two products of citrate are produced.
* This is considered to be a rate-limiting enzyme in the Krebs cycle.
*This step is not reversible.
*This enzyme is an induced fit enzyme (it goes into its closed form after the substrate binds to the active site).
The button above labeled backbone shows the backbone of this enzyme. Everything colored on blue would be considered the backbone. This is made out of chains of amino acids. This would be considered the primary structure of the enzyme.
The button above labeled Helix and Sheet shows the secondary structure of this enzyme. The secondary structure consists of the helix and the sheet. The helix is shown in purple and the sheet is shown in blue.
The button above called protein folding shows the tertiary structure that consists of protein folding. The protein is covered in green, and the citrate is colored white. Wireframe structure was used to try to show all of the folding.
1. Goodsell, D. (2007). Citrate Synthase. Retrieved from: http://pdb101.rcsb.org/motm/93
2. Kosmacki, A. Citrate Synthase. Retrieved from: https://collab.its.virginia.edu/access/content/group/f85bed6c-45d2-4b18-b868-6a2353586804/2/Ch19_Kosmacki_A_Citrate_Synthase_--Sus_scrofa-_-/Ch19_Kosmacki_A_Citrate_Synthase_--Sus_scrofa_Citrate_Synthase.html
3. https://pubchem.ncbi.nlm.nih.gov/compound/citrate#section=Top
4. (2016). Krebs Cycle/ TCA Cycle. Retrieved from: https://ramneetkaur.com/krebs-cycle-tca-cycle-mnemonic/