

This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
Penicillin was discovered through a chance observation in 1928 by Alexander Fleming.He discovered that colonies of Penicillium mold growing in a bacterial culture were able to stave off infection. A pond more investigation he found that the mold in the culture was producing penicillin that was killing off the bacteria. Penicillin is chemically similar to modular pieces that from the peptidoglycan. When penicillin is used as a drug, it blocks the enzyme (preclinical-binding proteins).
The secondary structure consist of 21% helical (17 helices; 158 residues) and 12% beta sheet (29 strands; 93 residues). Penicillin binding protein is 720 residues long which makes up a polypeptide(L).
PBPs are all involved in the final stages of the synthesis of peptidoglycan, which is the major component of bacterial cell walls. Bacterial cell wall synthesis is essential to growth, cell division (thus reproduction) and maintaining the cellular structure in bacteria. Inhibition of PBPs leads to irregularities in cell wall structure such as elongation, lesions, loss of selective permeability, and eventual cell death and lysis.
BP1b is a three-domain protein, comprising a
central transpeptidase domain flanked by a C-terminal β-strand-rich region and an N-terminal domain that links the transpeptidase and glycosyltransferase domains (latter domain mostly absent in the structure).
Depending on the structure and the catalytic activity of their N-terminal domain, they belong either to class A or class B PBPs. The C-terminal penicillin-binding domain of both classes has a transpeptidase (TP) activity catalyzing peptide cross-linking between two adjacent glycan chains. In class A, the N-terminal domain is responsible for their glycosyltransferase activity, catalyzing the elongation of uncross-linked glycan chains. In class B, the N-terminal domain is believed to play a role in cell morphogenesis by interacting with other proteins involved the cell cycle (Holtje, 1998). Monofunctional enzymes (MGTs) similar to the glycosyltransferase (GT) domain of class A PBPs (A-PBPs) also exist in some bacteria but their exact role is still unknown (Spratt, et al., 1996).
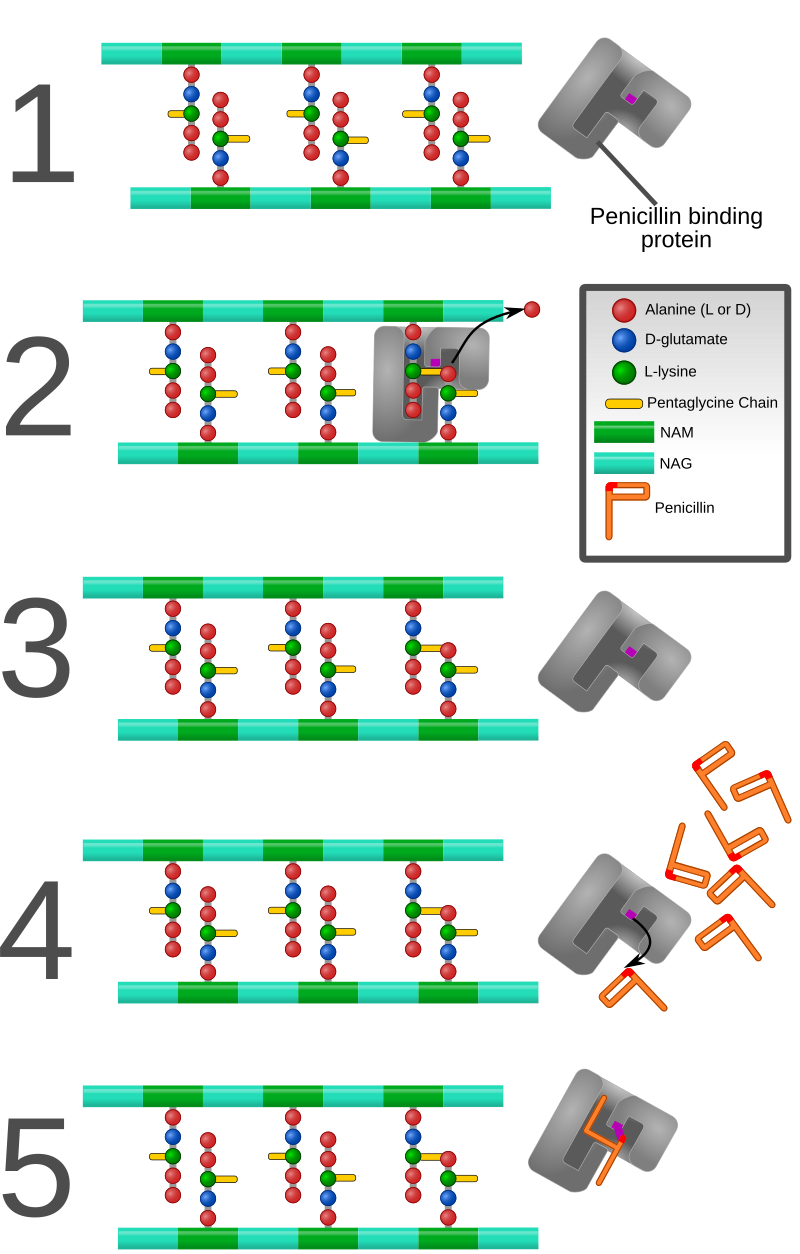
PBPs bind to β-lactam antibiotics because they are similar in chemical structure to the modular pieces that form the peptidoglycan. When they bind to penicillin, the β-lactam amide bond is ruptured to form a covalent bond with the catalytic serine residue at the PBPs active site. This is an irreversible reaction and inactivates the enzyme.
The PASTA domain is a small protein domain that can bind to beta-lactam rings found in antibiotics. The PASTA domain adopts a structure composed of an alpha-helix followed by three beta strands.
A ligand is an ion or molecule that binds to a central metal atom to form a complex.
Ligands



Remyfl. (2009, September 25). Penicillin Binding Protein Animation [Video file]. Retrieved from
https://www.youtube.com/watch?v=4uliYqX4j6M
Macheboeuf, P., Fische, D. S., Brown, T., Jr, Zervosen, A., Luxen, A., Joris,
B., Schofield, C. J. (2007). Structural and mechanistic basis of
penicillin-binding protein inhibition by lactivicins. Nature Chemical
Biology, (3), 565-569. http://dx.doi.org/10.1038/nchembio.2007.21