

This Jmol Exploration was created using the Jmol Exploration Webpage Creator from the MSOE Center for BioMolecular Modeling.
When exploring how a protein functions, we are often interested in seeing how the ligand interacts with the protein and what kinds of interactions these are.
An easy way to identify atoms that are within range to interact with a ligand is to use the following script. Any text in blue indicates a variable that must be specified based on the pdb file you are using. Using this script, the ligand you define will be colored in light cpk colors, which will make it easier to distinguish it from the interacting atoms.
Define ligbind (C41:O); Background white; Select all; backbone only; backbone off; Spacefill 1.25; Wireframe 1.0; Color cpk; Select ligbind and carbon; color [211, 211, 211]; Select ligbind and oxygen; color [255,185,185]; Select ligbind and nitrogen; color [160,210,255]; Center ligbind; Display ligbind; Contact{ligbind};
Display add contact(ligbind)
The following buttons use the drug aliskiren bound to the protein renin (2v0z.pdb) to demonstrate the script. The image can be rotated (click and drag the mouse on the image) or zoomed (shift click and drag the mouse) for different views of the protein. This first button shows the commands through 'Display ligbind'. With these commands, you are isolating the drug (ligand).
Display ligbindThe next button displays the script through 'contact{ligbind}' Here, you are displaying the nearest contacts of the drug. The disks indicate where the van der Waals radii of atoms overlaps. The colors indicate how close the contact is: yellow = close, orange = touching, and red = overlapping.
Contact{ligbind}The following button shows the whole script. Here, you are displaying the atoms that are in contact with your drug. These atoms are then flashed in cyan. (Flashing in cyan incorporates additional code, which is described further in the Jmol Animations Tutorial.)
Whole ScriptThe 'C41:O' in the script is modified to fit YOUR protein's ligand. You can find your protein's 3-character ligand code on pdb.org. In the example we use here, there are two identical chains in the protein, and the ':O' is identifying the ligand that is specifically on the O chain.
At this point, you will see all the atoms that are contacting your ligand, but you won't see the entire structure to which each of these atoms is attached. To see what these are, you'll need to hover your mouse over each atom that is displayed. (If your console is open, you may click on the atoms, and their identity will be displayed in the console.) Sometimes the nearest atoms are water molecules; these will appear as HOH and will only display the oxygen atoms. If you wish to display these, you will need to reference the water as HOH###, where ### represents the number following HOH on your screen.
Here are two atoms that are displayed as contacts to a ligand:
[HOH]2333:O.O #5983 36.075 15.323001 90.215
[ASP]215:O.OD1 #4298 37.652996 14.501 86.77
The first is a water molecule. Its name is HOH2333.
The second is an amino acid, asp215.
Make a list of all amino acids that interact with your ligand. (In this example, I've only chosen those that have touching or overlapping van der Waals radii — with orange or red in the center of the disk.) You'll need to display these — and determine whether sidechain or backbone atoms are interacting with the ligand. Use this script:
Define interact (:o and (thr12 or gln13 or tyr14 or asp32 or gly34 or ser76 or asp215 or hoh2333)); select contact(ligbind); spacefill off; wireframe off; display add interact; select interact or ligbind; wireframe 1.0; spacefill 1.25
Displaying Complete Amino AcidsThis will display complete amino acids on the :O chain, and then you can determine which part of the amino acid is interacting with the ligand. You can see that there are many small, yellow contacts on the drug, but these are not as significant as the orange- or red-colored contacts.
All amino acids share a common backbone structure and a sidechain (or R group) that varies from one amino acid to another. The identical backbones allow the amino acids to be joined together to form a peptide chain. The sidechains are attached to the 'alpha carbon' of the backbone. It is the order and interaction of these sidechains that gives each protein its unique characteristics.
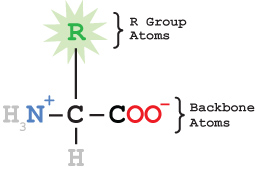
If your goal is to display the active site of a protein, or how the protein interacts with a ligand, you don't want to display all the atoms of the protein. Your image will simply be too cluttered. A common practice is to display the protein in an alpha carbon backbone (or backbone) format, which draws lines between adjacent alpha carbon atoms in the peptide chain. Then you display the ligand and any important interactions with the protein.
Most often, the interactions are with sidechain atoms which protrude from the protein backbone. Sometimes the interactions are with backbone atoms (typically oxygen or nitrogen atoms), and these need to be displayed using a different set of commands.
Therefore, you must identify whether the interactions with your ligand are with sidechain or backbone atoms of amino acids. We'll explore both types of interactions using the renin protein with the drug aliskiren (pdb file 2v0z). In each case, the amino acid sidechain flashes in cyan and the backbone flashes in lime green. The ligand (aliskiren) is in light cpk.
Sidechain InteractionYou can see how the backbone atoms of Arg74 are interacting with the drug...and the Arg74 sidechain itself isn't near the drug at all.
Backbone InteractionPlease note that the display commands, while extremely useful in exploring details of protein interactions, should be avoided in creating Jmol scripts for e-posters or models. Select and restrict commands are better in these applications.
Once you have determined what amino acids are interacting with your ligand, and whether the interacting atoms are backbone or sidechain atoms, you must decide what kind of interaction is occurring. Below are some example interactions that you may find in your protein. Click each button to see these interactions.
In each example, the amino acid(s) flash in cyan, water molecules or metal ions flash in purple (if shown), and the drug flashes in lime green (if shown).
Hydrogen bonding is displayed in renin with bound aliskiren, based on 2v0z.pdb. Note that the hydrogen atoms are not displayed in the structure, and at first glance, this may appear to be a charge interaction between the O- on the Asp sidechain (red) and the N+ on the ligand (light blue), but if you look at the ligand structure in the pdb file, you will see that the nitrogen is actually NH3+, which makes this a N-H...O interaction, which is a hydrogen bond interaction.
Hydrogen BondingAngiotensin converting enzyme with bound lisinopril (1o86.pdb) is used here. The interaction is between non-polar (hydrophobic) carbon atoms (grey) in the two structures.
Hydrophobic (van der Waals) InteractionLysozyme (1lpi.pdb) is used in the pi-cation interaction example. Note that the pi electrons typically are found in a donut shaped ring above and below a ring structure. The cation in this case is Na+.
Pi-Cation Interaction - Lysozyme PDB ID: 1lpiInteractions with a metal ion are shown in angiotensin converting enzyme with bound lisinopril (1o86.pdb). The metal is Zn2+, and it is coordinated (held in place) by a variety of interactions. Here the entire sidechain of the interacting amino acids flashes in cyan (to distinguish it from the backbone atoms), but the zinc interacts directly with the nearby oxygens (red) or nitrogens (blue). Only the interacting oxygens from lisinopril flash in lime green.
Metal Binding - ACE PDB ID: 1o86Here you see the stacking of two rings, which is characteristic of pi-pi interactions. This is angiotensin converting enzyme with lisinopril (1o86.pdb).
Pi-Pi Interaction - ACE PDB ID: 1o86This is renin with bound aliskiren, based on 2v0z.pdb. In this script, you first see sidechain oxygens of Gln128 and Thr295 flashing, followed by the oxygen on the drug. But the distance between these atoms is too great for a hydrogen bond. However, there is a water molecule in the crystal structure that serves as a bridge to faciliate hydrogen bonding between these atoms. After a brief pause, the water molecule appears, and then the interacting partners each flash color in turn - cyan for amino acids, purple for water, and lime green for the drug.
Water-Mediated Hydrogen Bonding - Renin PDB ID: 0v0zMany pdb files contain diagramatic sketches of ligand interactions in the Ligand Chemical Component section of the Summary tab. Although this may be a great place to begin exploring the interactions, we have found that many of these sketches omit some of the most significant interactions. To illustrate this point, we'll briefly explore the beta 1 adrenergic receptor with bound Albuterol (2y04.pdb). The first image is a screen shot of the PDB website, where you can find this contact image:
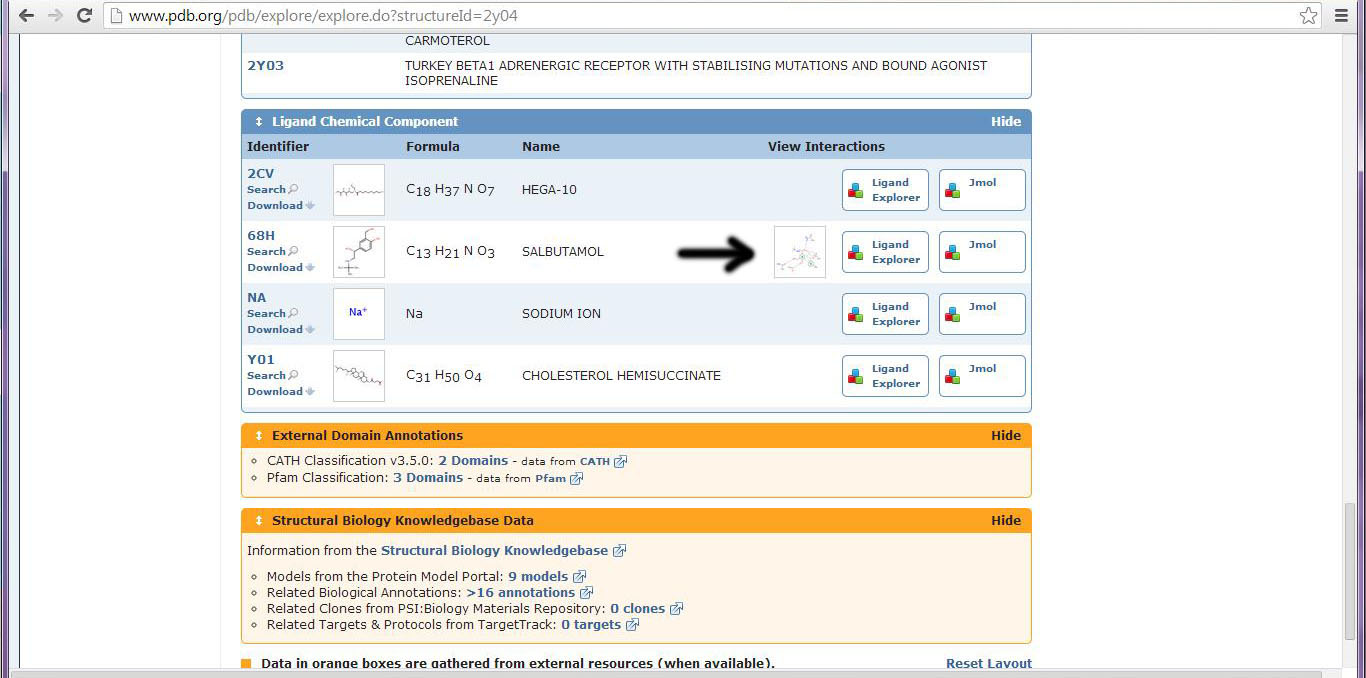
This is the contacts image from the pdb website for 2y04:
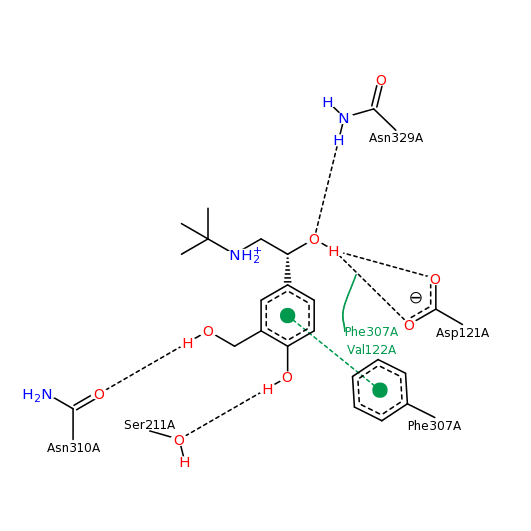
While these contacts are all correct, click the button below to see the contacts that Jmol calculates for Albuterol.
Contacts with Albuterol PDB ID: 2y04The flashing amino acid, Asp121, makes an extremely close contact with the nitrogen atom of Albuterol, as shown by the red-yellow disk between the two. This interaction, however, is not shown in the contact image from the PDB website. The primary citation associated with the pdb file confirms the interaction between Asp121 (D121) and the nitrogen atom of the Albuterol (Warne et al.,2011) .
Here is Figure 3 from the primary citation:
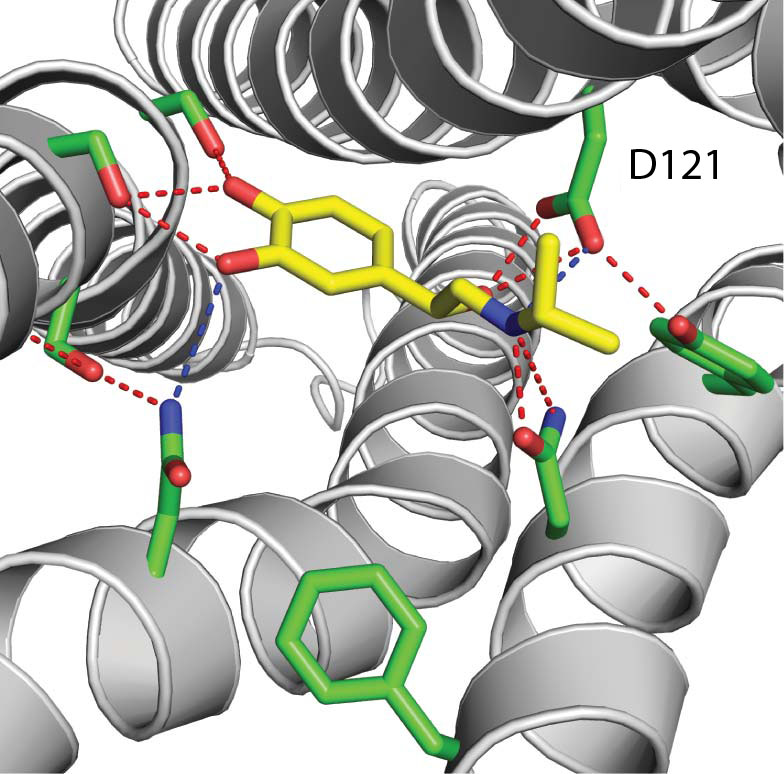
Although it may be easier to simply look at the contact image from the PDB website, exploring your protein with the 'contact' script may give you more insight into the interactions between your drug and your protein. We encourage you to consult the primary citation related to your pdb file in conjunction with exploring your structure in Jmol to identify the most relevant contacts.
Warne, T, Moukhametzianov, R, Baker, J.G., Nehmé, R., Edwards, P.C., Leslie, A.G., Schertler, G.F. and Tate, C.G. 2011. Structural basis for agonist and partial agonist action on a β(1)-adrenergic receptor. Nature 469(7329):241-244.